The novel Legionella pneumophila type II secretion substrate NttC contributes to infection of amoebae Hartmannella vermiformis and Willaertia magna
- PMID: 25253612
- PMCID: PMC4252911
- DOI: 10.1099/mic.0.082750-0
The novel Legionella pneumophila type II secretion substrate NttC contributes to infection of amoebae Hartmannella vermiformis and Willaertia magna
Abstract
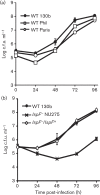
The type II protein secretion (T2S) system of Legionella pneumophila secretes over 25 proteins, including novel proteins that have no similarity to proteins of known function. T2S is also critical for the ability of L. pneumophila to grow within its natural amoebal hosts, including Acanthamoeba castellanii, Hartmannella vermiformis and Naegleria lovaniensis. Thus, T2S has an important role in the natural history of legionnaires' disease. Our previous work demonstrated that the novel T2S substrate NttA promotes intracellular infection of A. castellanii, whereas the secreted RNase SrnA, acyltransferase PlaC, and metalloprotease ProA all promote infection of H. vermiformis and N. lovaniensis. In this study, we determined that another novel T2S substrate that is specific to Legionella, designated NttC, is unique in being required for intracellular infection of H. vermiformis but not for infection of N. lovaniensis or A. castellanii. Expanding our repertoire of amoebal hosts, we determined that Willaertia magna is susceptible to infection by L. pneumophila strains 130b, Philadelphia-1 and Paris. Furthermore, T2S and, more specifically, NttA, NttC and PlaC were required for infection of W. magna. Taken together, these data demonstrate that the T2S system of L. pneumophila is critical for infection of at least four types of aquatic amoebae and that the importance of the individual T2S substrates varies in a host cell-specific fashion. Finally, it is now clear that novel T2S-dependent proteins that are specific to the genus Legionella are particularly important for L. pneumophila infection of key, environmental hosts.
© 2014 The Authors.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

