An agent-based model for mRNA export through the nuclear pore complex
- PMID: 25253717
- PMCID: PMC4230623
- DOI: 10.1091/mbc.E14-06-1065
An agent-based model for mRNA export through the nuclear pore complex
Abstract
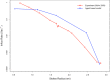
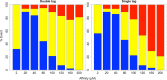
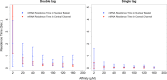
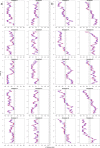
mRNA export from the nucleus is an essential step in the expression of every protein- coding gene in eukaryotes, but many aspects of this process remain poorly understood. The density of export receptors that must bind an mRNA to ensure export, as well as how receptor distribution affects transport dynamics, is not known. It is also unclear whether the rate-limiting step for transport occurs at the nuclear basket, in the central channel, or on the cytoplasmic face of the nuclear pore complex. Using previously published biophysical and biochemical parameters of mRNA export, we implemented a three-dimensional, coarse-grained, agent-based model of mRNA export in the nanosecond regime to gain insight into these issues. On running the model, we observed that mRNA export is sensitive to the number and distribution of transport receptors coating the mRNA and that there is a rate-limiting step in the nuclear basket that is potentially associated with the mRNA reconfiguring itself to thread into the central channel. Of note, our results also suggest that using a single location-monitoring mRNA label may be insufficient to correctly capture the time regime of mRNA threading through the pore and subsequent transport. This has implications for future experimental design to study mRNA transport dynamics.
© 2014 Azimi, Bulat, et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

