RNAi dynamics in Juvenile Fasciola spp. Liver flukes reveals the persistence of gene silencing in vitro
- PMID: 25254508
- PMCID: PMC4177864
- DOI: 10.1371/journal.pntd.0003185
RNAi dynamics in Juvenile Fasciola spp. Liver flukes reveals the persistence of gene silencing in vitro
Abstract
Background: Fasciola spp. liver fluke cause pernicious disease in humans and animals. Whilst current control is unsustainable due to anthelmintic resistance, gene silencing (RNA interference, RNAi) has the potential to contribute to functional validation of new therapeutic targets. The susceptibility of juvenile Fasciola hepatica to double stranded (ds)RNA-induced RNAi has been reported. To exploit this we probe RNAi dynamics, penetrance and persistence with the aim of building a robust platform for reverse genetics in liver fluke. We describe development of standardised RNAi protocols for a commercially-available liver fluke strain (the US Pacific North West Wild Strain), validated via robust transcriptional silencing of seven virulence genes, with in-depth experimental optimisation of three: cathepsin L (FheCatL) and B (FheCatB) cysteine proteases, and a σ-class glutathione transferase (FheσGST).
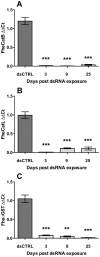
Methodology/principal findings: Robust transcriptional silencing of targets in both F. hepatica and Fasciola gigantica juveniles is achievable following exposure to long (200-320 nt) dsRNAs or 27 nt short interfering (si)RNAs. Although juveniles are highly RNAi-susceptible, they display slower transcript and protein knockdown dynamics than those reported previously. Knockdown was detectable following as little as 4h exposure to trigger (target-dependent) and in all cases silencing persisted for ≥25 days following long dsRNA exposure. Combinatorial silencing of three targets by mixing multiple long dsRNAs was similarly efficient. Despite profound transcriptional suppression, we found a significant time-lag before the occurrence of protein suppression; FheσGST and FheCatL protein suppression were only detectable after 9 and 21 days, respectively.
Conclusions/significance: In spite of marked variation in knockdown dynamics, we find that a transient exposure to long dsRNA or siRNA triggers robust RNAi penetrance and persistence in liver fluke NEJs supporting the development of multiple-throughput phenotypic screens for control target validation. RNAi persistence in fluke encourages in vivo studies on gene function using worms exposed to RNAi-triggers prior to infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

