Unusual influenza A viruses in bats
- PMID: 25256392
- PMCID: PMC4189031
- DOI: 10.3390/v6093438
Unusual influenza A viruses in bats
Abstract
Influenza A viruses infect a remarkably diverse number of hosts. Two completely new influenza A virus subtypes were recently discovered in bats, dramatically expanding the host range of the virus. These bat viruses are extremely divergent from all other known strains and likely have unique replication cycles. Phylogenetic analysis indicates long-term, isolated evolution in bats. This is supported by a high seroprevalence in sampled bat populations. As bats represent ~20% of all classified mammals, these findings suggests the presence of a massive cryptic reservoir of poorly characterized influenza A viruses. Here, we review the exciting progress made on understanding these newly discovered viruses, and discuss their zoonotic potential.
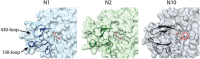
Figures

References
-
- Halpin K., Young P.L., Field H.E., Mackenzie J.S. Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol. 2000;81:1927–1932. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

