Oxidative stress and autophagy: the clash between damage and metabolic needs
- PMID: 25257172
- PMCID: PMC4326572
- DOI: 10.1038/cdd.2014.150
Oxidative stress and autophagy: the clash between damage and metabolic needs
Abstract
Autophagy is a catabolic process aimed at recycling cellular components and damaged organelles in response to diverse conditions of stress, such as nutrient deprivation, viral infection and genotoxic stress. A growing amount of evidence in recent years argues for oxidative stress acting as the converging point of these stimuli, with reactive oxygen species (ROS) and reactive nitrogen species (RNS) being among the main intracellular signal transducers sustaining autophagy. This review aims at providing novel insight into the regulatory pathways of autophagy in response to glucose and amino acid deprivation, as well as their tight interconnection with metabolic networks and redox homeostasis. The role of oxidative and nitrosative stress in autophagy is also discussed in the light of its being harmful for both cellular biomolecules and signal mediator through reversible posttranslational modifications of thiol-containing proteins. The redox-independent relationship between autophagy and antioxidant response, occurring through the p62/Keap1/Nrf2 pathway, is also addressed in order to provide a wide perspective upon the interconnection between autophagy and oxidative stress. Herein, we also attempt to afford an overview of the complex crosstalk between autophagy and DNA damage response (DDR), focusing on the main pathways activated upon ROS and RNS overproduction. Along these lines, the direct and indirect role of autophagy in DDR is dissected in depth.
Figures

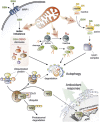
 ) and H2O2 are the main ROS produced by mitochondria upon nutrient deprivation. They positively regulate autophagy by means of at least three different mechanisms, including: (1) S-glutathionylation (SH → S-SG) of cysteines located in the α and β subunits of AMPK (top right); (2) oxidation of Cys81 (SH → Sox) of Atg4 that in turn leads to the inactivation of its ‘delipidating' activity on LC3 and to the accumulation of the pro-autophagic LC3-II isoform (top centre); and (3) wide alteration of thiol redox state (e.g., decrease of GSH/GSSG ratio and general increase of oxidized thiols, Sox) that is facilitated by the release of reduced glutathione (GSH) to the extracellular milieu through the multidrug resistance protein 1 (MRP1) (top left). In a redox-independent manner, it has also been demonstrated that p62, when bound to ubiquitylated protein aggregates, can undergo phosphorylation on Ser351, thereby sequestering Keap1 and leading to its detachment from Nrf2 (bottom left). Consequently, Nrf2 is no longer degraded by the ubiquitin-3 proteasome system, but translocates in the nucleus, binds to antioxidant-responsive elements (AREs) located in the promoter regions of antioxidant genes and activates their transcription (bottom right)
) and H2O2 are the main ROS produced by mitochondria upon nutrient deprivation. They positively regulate autophagy by means of at least three different mechanisms, including: (1) S-glutathionylation (SH → S-SG) of cysteines located in the α and β subunits of AMPK (top right); (2) oxidation of Cys81 (SH → Sox) of Atg4 that in turn leads to the inactivation of its ‘delipidating' activity on LC3 and to the accumulation of the pro-autophagic LC3-II isoform (top centre); and (3) wide alteration of thiol redox state (e.g., decrease of GSH/GSSG ratio and general increase of oxidized thiols, Sox) that is facilitated by the release of reduced glutathione (GSH) to the extracellular milieu through the multidrug resistance protein 1 (MRP1) (top left). In a redox-independent manner, it has also been demonstrated that p62, when bound to ubiquitylated protein aggregates, can undergo phosphorylation on Ser351, thereby sequestering Keap1 and leading to its detachment from Nrf2 (bottom left). Consequently, Nrf2 is no longer degraded by the ubiquitin-3 proteasome system, but translocates in the nucleus, binds to antioxidant-responsive elements (AREs) located in the promoter regions of antioxidant genes and activates their transcription (bottom right)

References
-
- De Duve C, Gianetto R, Appelmans F, Wattiaux R. Enzymic content of the mitochondria fraction. Nature. 1953;172:1143–1144. - PubMed
-
- De Duve C, Berthet J. Reproducibility of differential centrifugation experiments in tissue fractionation. Nature. 1953;172:1142. - PubMed
-
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. - PubMed
-
- De Duve C, Baudhuin P. Peroxisomes (microbodies and related particles) Physiol Rev. 1966;46:323–357. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

