Transcriptional diversity during lineage commitment of human blood progenitors
- PMID: 25258084
- PMCID: PMC4254742
- DOI: 10.1126/science.1251033
Transcriptional diversity during lineage commitment of human blood progenitors
Abstract
Blood cells derive from hematopoietic stem cells through stepwise fating events. To characterize gene expression programs driving lineage choice, we sequenced RNA from eight primary human hematopoietic progenitor populations representing the major myeloid commitment stages and the main lymphoid stage. We identified extensive cell type-specific expression changes: 6711 genes and 10,724 transcripts, enriched in non-protein-coding elements at early stages of differentiation. In addition, we found 7881 novel splice junctions and 2301 differentially used alternative splicing events, enriched in genes involved in regulatory processes. We demonstrated experimentally cell-specific isoform usage, identifying nuclear factor I/B (NFIB) as a regulator of megakaryocyte maturation-the platelet precursor. Our data highlight the complexity of fating events in closely related progenitor populations, the understanding of which is essential for the advancement of transplantation and regenerative medicine.
Copyright © 2014, American Association for the Advancement of Science.
Figures
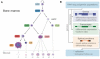
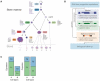
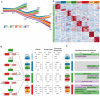
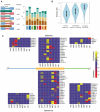

Comment in
-
Alternative splicing: Characterizing cell fate.Nat Rev Genet. 2014 Nov;15(11):706. doi: 10.1038/nrg3847. Nat Rev Genet. 2014. PMID: 25324005 No abstract available.
References
-
- Giebel B, Bruns I. Self-renewal versus differentiation in hematopoietic stem and progenitor cells: a focus on asymmetric cell divisions. Current stem cell research & therapy. 2008;3:9–16. doi. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 084183/Z/07/Z/WT_/Wellcome Trust/United Kingdom
- RG/09/012/28096/BHF_/British Heart Foundation/United Kingdom
- MR/K006584/1/MRC_/Medical Research Council/United Kingdom
- 082961/Z/07/Z/WT_/Wellcome Trust/United Kingdom
- 100140/WT_/Wellcome Trust/United Kingdom
- C45041/A14953/CRUK_/Cancer Research UK/United Kingdom
- 14953/CRUK_/Cancer Research UK/United Kingdom
- RP-PG-0310-1002/BHF_/British Heart Foundation/United Kingdom
- RP-PG-0310-1002/DH_/Department of Health/United Kingdom
- MR/K023489/1/MRC_/Medical Research Council/United Kingdom
- 082961/WT_/Wellcome Trust/United Kingdom
- WT098051/WT_/Wellcome Trust/United Kingdom
- MC_UP_0801/1/MRC_/Medical Research Council/United Kingdom
- MR/J011711/1/MRC_/Medical Research Council/United Kingdom
- FS/12/27/29405/BHF_/British Heart Foundation/United Kingdom
- 095908/WT_/Wellcome Trust/United Kingdom
- RG/09/12/28096/BHF_/British Heart Foundation/United Kingdom
- WT091310/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

