Detecting SNPs underlying domestication-related traits in soybean
- PMID: 25258093
- PMCID: PMC4180965
- DOI: 10.1186/s12870-014-0251-1
Detecting SNPs underlying domestication-related traits in soybean
Abstract
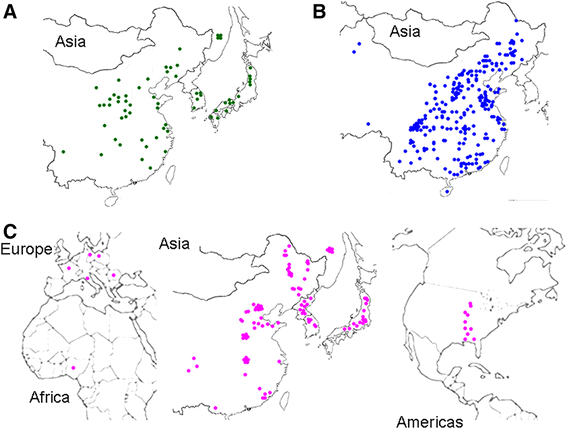
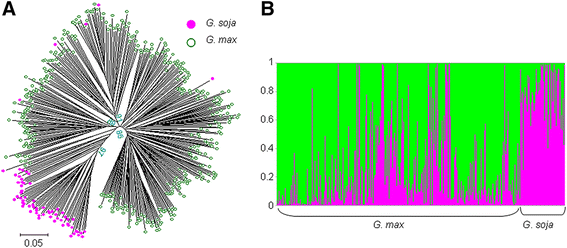
Background: Cultivated soybean (Glycine max) experienced a severe genetic bottleneck during its domestication and a further loss in diversity during its subsequent selection. Here, a panel of 65 wild (G. soja) and 353 cultivated accessions was genotyped at 552 single-nucleotide polymorphism loci to search for signals of selection during and after domestication.
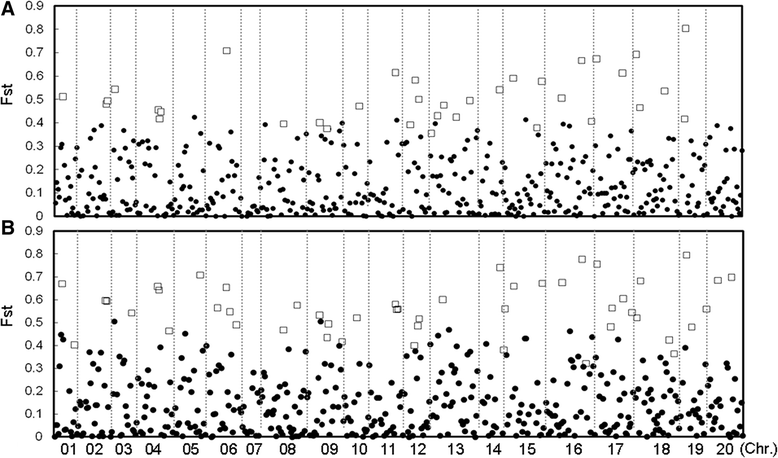
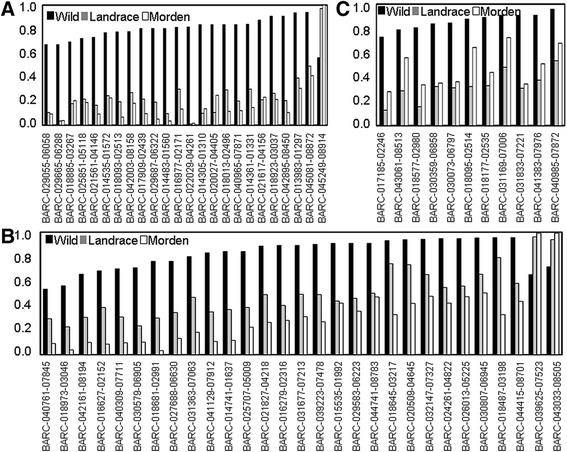
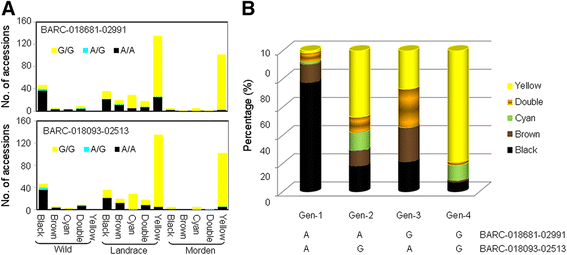
Results: The wild and cultivated populations were well differentiated from one another. Application of the Fst outlier test revealed 64 loci showing evidence for selection. Of these, 35 related to selection during domestication, while the other 29 likely gradually became monomorphic as a result of prolonged selection during post domestication. Two of the SNP locus outliers were associated with testa color.
Conclusions: Identifying genes controlling domestication-related traits is important for maintaining the diversity of crops. SNP locus outliers detected by a combined forward genetics and population genetics approach can provide markers with utility for the conservation of wild accessions and for trait improvement in the cultivated genepool.
Figures
Similar articles
-
Genetic diversity in domesticated soybean (Glycine max) and its wild progenitor (Glycine soja) for simple sequence repeat and single-nucleotide polymorphism loci.New Phytol. 2010 Oct;188(1):242-53. doi: 10.1111/j.1469-8137.2010.03344.x. Epub 2010 Jul 6. New Phytol. 2010. PMID: 20618914
-
Domestication footprints anchor genomic regions of agronomic importance in soybeans.New Phytol. 2016 Jan;209(2):871-84. doi: 10.1111/nph.13626. Epub 2015 Oct 19. New Phytol. 2016. PMID: 26479264
-
Genetic dissection of yield-related traits via genome-wide association analysis across multiple environments in wild soybean (Glycine soja Sieb. and Zucc.).Planta. 2020 Jan 6;251(2):39. doi: 10.1007/s00425-019-03329-6. Planta. 2020. PMID: 31907621
-
[Occurrence characteristics and molecular genetic basis of pod shattering in soybean].Yi Chuan. 2015 Jun;37(6):535-43. doi: 10.16288/j.yczz.14-456. Yi Chuan. 2015. PMID: 26351049 Review. Chinese.
-
Photoperiodism dynamics during the domestication and improvement of soybean.Sci China Life Sci. 2017 Dec;60(12):1416-1427. doi: 10.1007/s11427-016-9154-x. Epub 2017 Sep 21. Sci China Life Sci. 2017. PMID: 28942538 Review.
Cited by
-
Population structure and genetic diversity characterization of soybean for seed longevity.PLoS One. 2022 Dec 6;17(12):e0278631. doi: 10.1371/journal.pone.0278631. eCollection 2022. PLoS One. 2022. PMID: 36472991 Free PMC article.
-
Neurogenomics and the role of a large mutational target on rapid behavioral change.Biol Direct. 2016 Nov 8;11(1):60. doi: 10.1186/s13062-016-0162-1. Biol Direct. 2016. PMID: 27825385 Free PMC article.
-
Molecular phylogeny and dynamic evolution of disease resistance genes in the legume family.BMC Genomics. 2016 May 26;17:402. doi: 10.1186/s12864-016-2736-9. BMC Genomics. 2016. PMID: 27229309 Free PMC article.
-
Limited genetic diversity found among genotypes of the Entada landrace (Ensete ventricosum, (Welw.) Chessman) from Ethiopia.Front Plant Sci. 2024 Sep 9;15:1336461. doi: 10.3389/fpls.2024.1336461. eCollection 2024. Front Plant Sci. 2024. PMID: 39315368 Free PMC article.
-
SNP identification and marker assay development for high-throughput selection of soybean cyst nematode resistance.BMC Genomics. 2015 Apr 18;16(1):314. doi: 10.1186/s12864-015-1531-3. BMC Genomics. 2015. PMID: 25903750 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

