Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis
- PMID: 25258143
- PMCID: PMC4253562
- DOI: 10.1016/j.jaci.2014.07.021
Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis
Abstract
Background: Eosinophilic esophagitis (EoE) is a chronic antigen-driven allergic inflammatory disease, likely involving the interplay of genetic and environmental factors, yet their respective contributions to heritability are unknown.
Objective: To quantify the risk associated with genes and environment on familial clustering of EoE.
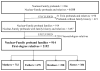
Methods: Family history was obtained from a hospital-based cohort of 914 EoE probands (n = 2192 first-degree "Nuclear-Family" relatives) and an international registry of monozygotic and dizygotic twins/triplets (n = 63 EoE "Twins" probands). Frequencies, recurrence risk ratios (RRRs), heritability, and twin concordance were estimated. Environmental exposures were preliminarily examined.
Results: Analysis of the Nuclear-Family-based cohort revealed that the rate of EoE, in first-degree relatives of a proband, was 1.8% (unadjusted) and 2.3% (sex-adjusted). RRRs ranged from 10 to 64, depending on the family relationship, and were higher in brothers (64.0; P = .04), fathers (42.9; P = .004), and males (50.7; P < .001) than in sisters, mothers, and females, respectively. The risk of EoE for other siblings was 2.4%. In the Nuclear-Family cohort, combined gene and common environment heritability was 72.0% ± 2.7% (P < .001). In the Twins cohort, genetic heritability was 14.5% ± 4.0% (P < .001), and common family environment contributed 81.0% ± 4% (P < .001) to phenotypic variance. Probandwise concordance in monozygotic co-twins was 57.9% ± 9.5% compared with 36.4% ± 9.3% in dizygotic co-twins (P = .11). Greater birth weight difference between twins (P = .01), breast-feeding (P = .15), and fall birth season (P = .02) were associated with twin discordance in disease status.
Conclusions: EoE RRRs are increased 10- to 64-fold compared with the general population. EoE in relatives is 1.8% to 2.4%, depending on relationship and sex. Nuclear-Family heritability appeared to be high (72.0%). However, the Twins cohort analysis revealed a powerful role for common environment (81.0%) compared with additive genetic heritability (14.5%).
Keywords: Eosinophilia; drug hypersensitivity; food allergy; gastrointestinal diseases; gene-environment interaction; heritability; immune system diseases; medical genetics; skin diseases; twins.
Copyright © 2014 American Academy of Allergy, Asthma & Immunology. All rights reserved.
Figures







References
-
- Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004 Aug 26;351(9):940–1. - PubMed
-
- Sealock RJ, Rendon G, El-Serag HB. Systematic review: The epidemiology of eosinophilic oesophagitis in adults. Aliment Pharmacol Ther. 2010 Sep;32(6):712–9. - PubMed
-
- Soon IS, Butzner JD, Kaplan GG, Debruyn JC. Incidence and prevalence of eosinophilic esophagitis in children: Systematic review and meta-analysis. J Pediatr Gastroenterol Nutr. 2013 Jul;57(1):72–80. - PubMed
Publication types
MeSH terms
Grants and funding
- 1K24DK100303/DK/NIDDK NIH HHS/United States
- UL1-TR000077-04/TR/NCATS NIH HHS/United States
- 1R25GM093044-01/GM/NIGMS NIH HHS/United States
- UL1-RR026314-01/RR/NCRR NIH HHS/United States
- P30 ES006096/ES/NIEHS NIH HHS/United States
- K24 DK100303/DK/NIDDK NIH HHS/United States
- UL1 TR000077/TR/NCATS NIH HHS/United States
- T32 ES010957/ES/NIEHS NIH HHS/United States
- UL1 RR026314/RR/NCRR NIH HHS/United States
- T32-ES10957/ES/NIEHS NIH HHS/United States
- P30 DK078392/DK/NIDDK NIH HHS/United States
- P30-ES006096/ES/NIEHS NIH HHS/United States
- R25 GM093044/GM/NIGMS NIH HHS/United States
- UL1 TR001425/TR/NCATS NIH HHS/United States
- I01 BX001834/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

