Torque generation of Enterococcus hirae V-ATPase
- PMID: 25258315
- PMCID: PMC4223323
- DOI: 10.1074/jbc.M114.598177
Torque generation of Enterococcus hirae V-ATPase
Abstract
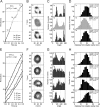
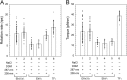
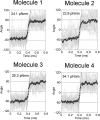
V-ATPase (V(o)V1) converts the chemical free energy of ATP into an ion-motive force across the cell membrane via mechanical rotation. This energy conversion requires proper interactions between the rotor and stator in V(o)V1 for tight coupling among chemical reaction, torque generation, and ion transport. We developed an Escherichia coli expression system for Enterococcus hirae V(o)V1 (EhV(o)V1) and established a single-molecule rotation assay to measure the torque generated. Recombinant and native EhV(o)V1 exhibited almost identical dependence of ATP hydrolysis activity on sodium ion and ATP concentrations, indicating their functional equivalence. In a single-molecule rotation assay with a low load probe at high ATP concentration, EhV(o)V1 only showed the "clear" state without apparent backward steps, whereas EhV1 showed two states, "clear" and "unclear." Furthermore, EhV(o)V1 showed slower rotation than EhV1 without the three distinct pauses separated by 120° that were observed in EhV1. When using a large probe, EhV(o)V1 showed faster rotation than EhV1, and the torque of EhV(o)V1 estimated from the continuous rotation was nearly double that of EhV1. On the other hand, stepping torque of EhV1 in the clear state was comparable with that of EhV(o)V1. These results indicate that rotor-stator interactions of the V(o) moiety and/or sodium ion transport limit the rotation driven by the V1 moiety, and the rotor-stator interactions in EhV(o)V1 are stabilized by two peripheral stalks to generate a larger torque than that of isolated EhV1. However, the torque value was substantially lower than that of other rotary ATPases, implying the low energy conversion efficiency of EhV(o)V1.
Keywords: Bioenergetics; Membrane Protein; Molecular Motor; Single-molecule Biophysics; V-ATPase; Vacuolar ATPase.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Nishi T., Forgac M. (2002) The vacuolar H+-ATPases: nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3, 94–103 - PubMed
-
- Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 - PubMed
-
- Bowman E. J., Bowman B. J. (2005) V-ATPases as drug targets. J. Bioenerg. Biomembr. 37, 431–435 - PubMed
-
- Stewart A. G., Laming E. M., Sobti M., Stock D. (2014) Rotary ATPases: dynamic molecular machines. Curr. Opin. Struct. Biol. 25C, 40–48 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

