A foundation for provitamin A biofortification of maize: genome-wide association and genomic prediction models of carotenoid levels
- PMID: 25258377
- PMCID: PMC4256781
- DOI: 10.1534/genetics.114.169979
A foundation for provitamin A biofortification of maize: genome-wide association and genomic prediction models of carotenoid levels
Abstract
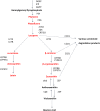
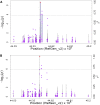
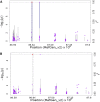
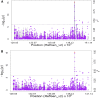
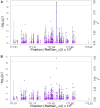
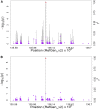
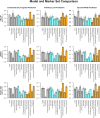
Efforts are underway for development of crops with improved levels of provitamin A carotenoids to help combat dietary vitamin A deficiency. As a global staple crop with considerable variation in kernel carotenoid composition, maize (Zea mays L.) could have a widespread impact. We performed a genome-wide association study (GWAS) of quantified seed carotenoids across a panel of maize inbreds ranging from light yellow to dark orange in grain color to identify some of the key genes controlling maize grain carotenoid composition. Significant associations at the genome-wide level were detected within the coding regions of zep1 and lut1, carotenoid biosynthetic genes not previously shown to impact grain carotenoid composition in association studies, as well as within previously associated lcyE and crtRB1 genes. We leveraged existing biochemical and genomic information to identify 58 a priori candidate genes relevant to the biosynthesis and retention of carotenoids in maize to test in a pathway-level analysis. This revealed dxs2 and lut5, genes not previously associated with kernel carotenoids. In genomic prediction models, use of markers that targeted a small set of quantitative trait loci associated with carotenoid levels in prior linkage studies were as effective as genome-wide markers for predicting carotenoid traits. Based on GWAS, pathway-level analysis, and genomic prediction studies, we outline a flexible strategy involving use of a small number of genes that can be selected for rapid conversion of elite white grain germplasm, with minimal amounts of carotenoids, to orange grain versions containing high levels of provitamin A.
Keywords: biofortification; carotenoid; genome-wide association study; genomic prediction; pathway-level analysis; provitamin A.
Copyright © 2014 by the Genetics Society of America.
Figures
Similar articles
-
Genetic Loci Controlling Carotenoid Biosynthesis in Diverse Tropical Maize Lines.G3 (Bethesda). 2018 Mar 2;8(3):1049-1065. doi: 10.1534/g3.117.300511. G3 (Bethesda). 2018. PMID: 29378820 Free PMC article.
-
Genome-Wide Association Study and Pathway-Level Analysis of Kernel Color in Maize.G3 (Bethesda). 2019 Jun 5;9(6):1945-1955. doi: 10.1534/g3.119.400040. G3 (Bethesda). 2019. PMID: 31010822 Free PMC article.
-
Investigating genomic prediction strategies for grain carotenoid traits in a tropical/subtropical maize panel.G3 (Bethesda). 2024 May 7;14(5):jkae044. doi: 10.1093/g3journal/jkae044. G3 (Bethesda). 2024. PMID: 38427914 Free PMC article.
-
The development and release of maize fortified with provitamin A carotenoids in developing countries.Crit Rev Food Sci Nutr. 2019;59(8):1284-1293. doi: 10.1080/10408398.2017.1402751. Epub 2017 Dec 4. Crit Rev Food Sci Nutr. 2019. PMID: 29200311 Review.
-
Carotenoids in Staple Cereals: Metabolism, Regulation, and Genetic Manipulation.Front Plant Sci. 2016 Aug 10;7:1197. doi: 10.3389/fpls.2016.01197. eCollection 2016. Front Plant Sci. 2016. PMID: 27559339 Free PMC article. Review.
Cited by
-
Genome-wide association analysis reveals new targets for carotenoid biofortification in maize.Theor Appl Genet. 2015 May;128(5):851-64. doi: 10.1007/s00122-015-2475-3. Epub 2015 Feb 18. Theor Appl Genet. 2015. PMID: 25690716 Free PMC article.
-
Phenotypic characterization and validation of provitamin A functional genes in early maturing provitamin A-quality protein maize (Zea mays) inbred lines.Plant Breed. 2020 Jun;139(3):575-588. doi: 10.1111/pbr.12798. Epub 2019 Dec 20. Plant Breed. 2020. PMID: 32742048 Free PMC article.
-
The utility of metabolomics as a tool to inform maize biology.Plant Commun. 2021 Apr 21;2(4):100187. doi: 10.1016/j.xplc.2021.100187. eCollection 2021 Jul 12. Plant Commun. 2021. PMID: 34327322 Free PMC article. Review.
-
Verification of Three-Phase Dependency Analysis Bayesian Network Learning Method for Maize Carotenoid Gene Mining.Biomed Res Int. 2017;2017:1813494. doi: 10.1155/2017/1813494. Epub 2017 Jul 30. Biomed Res Int. 2017. PMID: 28828382 Free PMC article.
-
Genome assembly and population genomic analysis reveal the genetic basis of popcorn evolution.Plant Biotechnol J. 2025 Jul;23(7):2911-2927. doi: 10.1111/pbi.70125. Epub 2025 May 5. Plant Biotechnol J. 2025. PMID: 40322841 Free PMC article.
References
-
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

