Widespread intron retention in mammals functionally tunes transcriptomes
- PMID: 25258385
- PMCID: PMC4216919
- DOI: 10.1101/gr.177790.114
Widespread intron retention in mammals functionally tunes transcriptomes
Abstract
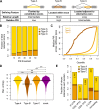
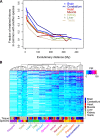
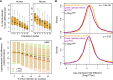
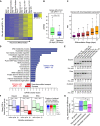
Alternative splicing (AS) of precursor RNAs is responsible for greatly expanding the regulatory and functional capacity of eukaryotic genomes. Of the different classes of AS, intron retention (IR) is the least well understood. In plants and unicellular eukaryotes, IR is the most common form of AS, whereas in animals, it is thought to represent the least prevalent form. Using high-coverage poly(A)(+) RNA-seq data, we observe that IR is surprisingly frequent in mammals, affecting transcripts from as many as three-quarters of multiexonic genes. A highly correlated set of cis features comprising an "IR code" reliably discriminates retained from constitutively spliced introns. We show that IR acts widely to reduce the levels of transcripts that are less or not required for the physiology of the cell or tissue type in which they are detected. This "transcriptome tuning" function of IR acts through both nonsense-mediated mRNA decay and nuclear sequestration and turnover of IR transcripts. We further show that IR is linked to a cross-talk mechanism involving localized stalling of RNA polymerase II (Pol II) and reduced availability of spliceosomal components. Collectively, the results implicate a global checkpoint-type mechanism whereby reduced recruitment of splicing components coupled to Pol II pausing underlies widespread IR-mediated suppression of inappropriately expressed transcripts.
© 2014 Braunschweig et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B, et al. 2012. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep 1: 543–556 - PubMed
-
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593 - PubMed
-
- Beckers JM, Rixen M. 2003. EOF calculations and data filling from incomplete oceanographic datasets. J Atmos Ocean Technol 20: 1839–1856
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
