Modeling of human M1 aminopeptidases for in silico screening of potential Plasmodium falciparum alanine aminopeptidase (PfA-M1) specific inhibitors
- PMID: 25258488
- PMCID: PMC4166772
- DOI: 10.6026/97320630010518
Modeling of human M1 aminopeptidases for in silico screening of potential Plasmodium falciparum alanine aminopeptidase (PfA-M1) specific inhibitors
Abstract
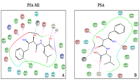
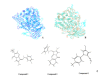
Plasmodium falciparum alanine M1-aminopeptidase (PfA-M1) is a validated target for anti-malarial drug development. Presence of significant similarity between PfA-M1 and human M1-aminopeptidases, particularly within regions of enzyme active site leads to problem of non-specificity and off-target binding for known aminopeptidase inhibitors. Molecular docking based in silico screening approach for off-target binding has high potential but requires 3D-structure of all human M1-aminopeptidaes. Therefore, in the present study 3D structural models of seven human M1-aminopeptidases were developed. The robustness of docking parameters and quality of predicted human M1-aminopeptidases structural models was evaluated by stereochemical analysis and docking of their respective known inhibitors. The docking scores were in agreement with the inhibitory concentrations elucidated in enzyme assays of respective inhibitor enzyme combinations (r2≈0.70). Further docking analysis of fifteen potential PfA-M1 inhibitors (virtual screening identified) showed that three compounds had less docking affinity for human M1-aminopeptidases as compared to PfA-M1. These three identified potential lead compounds can be validated with enzyme assays and used as a scaffold for designing of new compounds with increased specificity towards PfA-M1.
Keywords: Drug designing; homology modeling; in silico screening; malaria; molecular docking.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
