Comparative genomics of planktonic Flavobacteriaceae from the Gulf of Maine using metagenomic data
- PMID: 25258679
- PMCID: PMC4164334
- DOI: 10.1186/2049-2618-2-34
Comparative genomics of planktonic Flavobacteriaceae from the Gulf of Maine using metagenomic data
Abstract
Background: The Gulf of Maine is an important biological province of the Northwest Atlantic with high productivity year round. From an environmental Sanger-based metagenome, sampled in summer and winter, we were able to assemble and explore the partial environmental genomes of uncultured members of the class Flavobacteria. Each of the environmental genomes represents organisms that compose less than 1% of the total microbial metagenome.
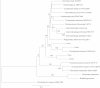
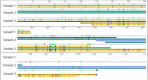
Results: Four partial environmental genomes were assembled with varying degrees of estimated completeness (37%-84% complete) and were analyzed from a perspective of gathering information regarding niche partitioning between co-occurring organisms. Comparative genomics revealed potentially important niche partitioning genomic variations, including iron transporters and genes associated with cell attachment and polymer degradation. Analysis of large syntenic regions helped reveal potentially ecologically relevant variations for Flavobacteriaceae in the Gulf of Maine, such as arginine biosynthesis, and identify a putative genomic island incorporating novel exogenous genes from the environment.
Conclusions: Biogeographic analysis revealed flavobacteria species with distinct abundance patterns suggesting the presence of local blooms relative to the other species, as well as seasonally selected organisms. The analysis of genomic content for the Gulf of Maine Flavobacteria supports the hypothesis of a particle-associated lifestyle and specifically highlights a number of putative coding sequences that may play a role in the remineralization of particulate organic matter. And lastly, analysis of the underlying sequences for each assembled genome revealed seasonal and nonseasonal variants of specific genes implicating a dynamic interaction between individuals within the species.
Keywords: Comparative genomics; Metagenomics; Microbial ecology.
Figures



Similar articles
-
Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine.Environ Microbiol. 2012 Jan;14(1):254-67. doi: 10.1111/j.1462-2920.2011.02628.x. Epub 2011 Nov 3. Environ Microbiol. 2012. PMID: 22050608
-
Assembling the marine metagenome, one cell at a time.PLoS One. 2009;4(4):e5299. doi: 10.1371/journal.pone.0005299. Epub 2009 Apr 23. PLoS One. 2009. PMID: 19390573 Free PMC article.
-
Genomic Characterization of Candidate Division LCP-89 Reveals an Atypical Cell Wall Structure, Microcompartment Production, and Dual Respiratory and Fermentative Capacities.Appl Environ Microbiol. 2019 May 2;85(10):e00110-19. doi: 10.1128/AEM.00110-19. Print 2019 May 15. Appl Environ Microbiol. 2019. PMID: 30902854 Free PMC article.
-
Metagenomics: application of genomics to uncultured microorganisms.Microbiol Mol Biol Rev. 2004 Dec;68(4):669-85. doi: 10.1128/MMBR.68.4.669-685.2004. Microbiol Mol Biol Rev. 2004. PMID: 15590779 Free PMC article. Review.
-
From cultured to uncultured genome sequences: metagenomics and modeling microbial ecosystems.Cell Mol Life Sci. 2015 Nov;72(22):4287-308. doi: 10.1007/s00018-015-2004-1. Epub 2015 Aug 9. Cell Mol Life Sci. 2015. PMID: 26254872 Free PMC article. Review.
Cited by
-
Microdiversity Shapes the Seasonal Niche of Prokaryotic Plankton Inhabiting Surface Waters in a Coastal Upwelling System.Environ Microbiol Rep. 2025 Aug;17(4):e70131. doi: 10.1111/1758-2229.70131. Environ Microbiol Rep. 2025. PMID: 40692173 Free PMC article.
-
Identification of closely related species in Aspergillus through Analysis of Whole-Genome.Front Microbiol. 2024 Feb 21;15:1323572. doi: 10.3389/fmicb.2024.1323572. eCollection 2024. Front Microbiol. 2024. PMID: 38450170 Free PMC article.
-
Distributions of Extracellular Peptidases Across Prokaryotic Genomes Reflect Phylogeny and Habitat.Front Microbiol. 2019 Mar 5;10:413. doi: 10.3389/fmicb.2019.00413. eCollection 2019. Front Microbiol. 2019. PMID: 30891022 Free PMC article.
-
Variability in metagenomic samples from the Puget Sound: Relationship to temporal and anthropogenic impacts.PLoS One. 2018 Feb 13;13(2):e0192412. doi: 10.1371/journal.pone.0192412. eCollection 2018. PLoS One. 2018. PMID: 29438385 Free PMC article.
-
Post-translational modifications are enriched within protein functional groups important to bacterial adaptation within a deep-sea hydrothermal vent environment.Microbiome. 2016 Sep 6;4(1):49. doi: 10.1186/s40168-016-0194-x. Microbiome. 2016. PMID: 27600525 Free PMC article.
References
-
- Kirchman DL. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol. 2002;39:91–100. - PubMed
-
- Fuhrman JA, Azam F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar Biol. 1982;66:109–120. doi: 10.1007/BF00397184. - DOI
-
- Martinez J, Smith DC, Steward GF, Azam F. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat Microb Ecol. 1996;10:223–230.
LinkOut - more resources
Full Text Sources
Other Literature Sources

