Contrasting nitrogen fertilization treatments impact xylem gene expression and secondary cell wall lignification in Eucalyptus
- PMID: 25260963
- PMCID: PMC4189757
- DOI: 10.1186/s12870-014-0256-9
Contrasting nitrogen fertilization treatments impact xylem gene expression and secondary cell wall lignification in Eucalyptus
Abstract
Background: Nitrogen (N) is a main nutrient required for tree growth and biomass accumulation. In this study, we analyzed the effects of contrasting nitrogen fertilization treatments on the phenotypes of fast growing Eucalyptus hybrids (E. urophylla x E. grandis) with a special focus on xylem secondary cell walls and global gene expression patterns.
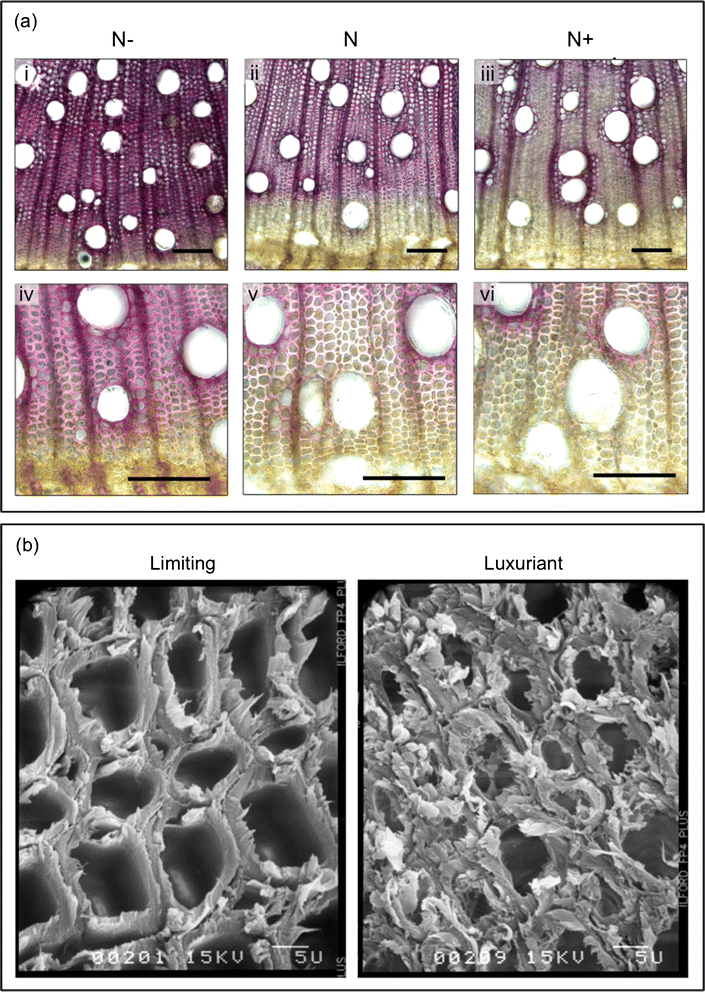
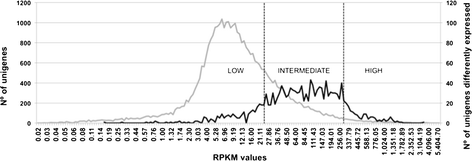
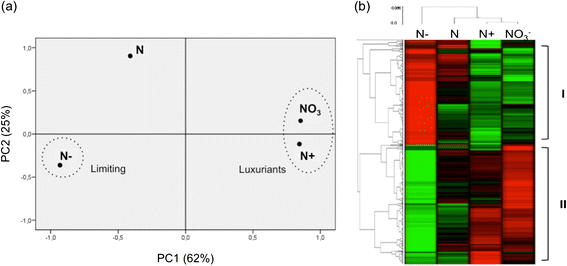
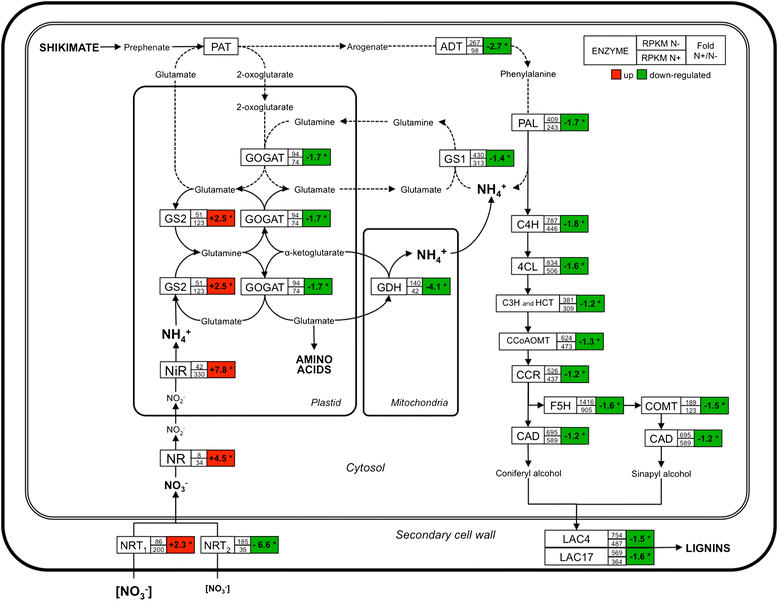
Results: Histological observations of the xylem secondary cell walls further confirmed by chemical analyses showed that lignin was reduced by luxuriant fertilization, whereas a consistent lignin deposition was observed in trees grown in N-limiting conditions. Also, the syringyl/guaiacyl (S/G) ratio was significantly lower in luxuriant nitrogen samples. Deep sequencing RNAseq analyses allowed us to identify a high number of differentially expressed genes (1,469) between contrasting N treatments. This number is dramatically higher than those obtained in similar studies performed in poplar but using microarrays. Remarkably, all the genes involved the general phenylpropanoid metabolism and lignin pathway were found to be down-regulated in response to high N availability. These findings further confirmed by RT-qPCR are in agreement with the reduced amount of lignin in xylem secondary cell walls of these plants.
Conclusions: This work enabled us to identify, at the whole genome level, xylem genes differentially regulated by N availability, some of which are involved in the environmental control of xylogenesis. It further illustrates that N fertilization can be used to alter the quantity and quality of lignocellulosic biomass in Eucalyptus, offering exciting prospects for the pulp and paper industry and for the use of short coppices plantations to produce second generation biofuels.
Figures



Similar articles
-
Flavonoid supplementation affects the expression of genes involved in cell wall formation and lignification metabolism and increases sugar content and saccharification in the fast-growing eucalyptus hybrid E. urophylla x E. grandis.BMC Plant Biol. 2014 Nov 19;14:301. doi: 10.1186/s12870-014-0301-8. BMC Plant Biol. 2014. PMID: 25407319 Free PMC article.
-
Eucalyptus hairy roots, a fast, efficient and versatile tool to explore function and expression of genes involved in wood formation.Plant Biotechnol J. 2016 Jun;14(6):1381-93. doi: 10.1111/pbi.12502. Epub 2015 Nov 18. Plant Biotechnol J. 2016. PMID: 26579999 Free PMC article.
-
The Eucalyptus linker histone variant EgH1.3 cooperates with the transcription factor EgMYB1 to control lignin biosynthesis during wood formation.New Phytol. 2017 Jan;213(1):287-299. doi: 10.1111/nph.14129. Epub 2016 Aug 8. New Phytol. 2017. PMID: 27500520
-
A review of xylan and lignin biosynthesis: foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes.Crit Rev Biochem Mol Biol. 2014 May-Jun;49(3):212-41. doi: 10.3109/10409238.2014.889651. Epub 2014 Feb 24. Crit Rev Biochem Mol Biol. 2014. PMID: 24564339 Review.
-
Woody plant cell walls: Fundamentals and utilization.Mol Plant. 2024 Jan 1;17(1):112-140. doi: 10.1016/j.molp.2023.12.008. Epub 2023 Dec 15. Mol Plant. 2024. PMID: 38102833 Review.
Cited by
-
Plant mineral nutrition and disease resistance: A significant linkage for sustainable crop protection.Front Plant Sci. 2022 Oct 20;13:883970. doi: 10.3389/fpls.2022.883970. eCollection 2022. Front Plant Sci. 2022. PMID: 36340341 Free PMC article. Review.
-
The Genetic Architecture of Nitrogen Use Efficiency in Switchgrass (Panicum virgatum L.).Front Plant Sci. 2022 May 2;13:893610. doi: 10.3389/fpls.2022.893610. eCollection 2022. Front Plant Sci. 2022. PMID: 35586220 Free PMC article.
-
Nitrogen deficiency results in changes to cell wall composition of sorghum seedlings.Sci Rep. 2021 Dec 2;11(1):23309. doi: 10.1038/s41598-021-02570-y. Sci Rep. 2021. PMID: 34857783 Free PMC article.
-
Network Analysis of Metabolome and Transcriptome Revealed Regulation of Different Nitrogen Concentrations on Hybrid Poplar Cambium Development.Int J Mol Sci. 2024 Jan 13;25(2):1017. doi: 10.3390/ijms25021017. Int J Mol Sci. 2024. PMID: 38256092 Free PMC article.
-
Effect of Nickel Ions on the Physiological and Transcriptional Responses to Carbon and Nitrogen Metabolism in Tomato Roots under Low Nitrogen Levels.Int J Mol Sci. 2022 Sep 27;23(19):11398. doi: 10.3390/ijms231911398. Int J Mol Sci. 2022. PMID: 36232700 Free PMC article.
References
-
- Séguin A. How could forest trees play an important role as feedstock for bioenergy production? Curr Opin Environ Sustain. 2011;3:90–94. doi: 10.1016/j.cosust.2010.12.006. - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

