Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features
- PMID: 25261934
- PMCID: PMC4343211
- DOI: 10.1038/ng.3103
Mutations in SPRTN cause early onset hepatocellular carcinoma, genomic instability and progeroid features
Abstract
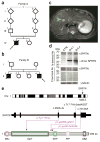
Age-related degenerative and malignant diseases represent major challenges for health care systems. Elucidation of the molecular mechanisms underlying carcinogenesis and age-associated pathologies is thus of growing biomedical relevance. We identified biallelic germline mutations in SPRTN (also called C1orf124 or DVC1) in three patients from two unrelated families. All three patients are affected by a new segmental progeroid syndrome characterized by genomic instability and susceptibility toward early onset hepatocellular carcinoma. SPRTN was recently proposed to have a function in translesional DNA synthesis and the prevention of mutagenesis. Our in vivo and in vitro characterization of identified mutations has uncovered an essential role for SPRTN in the prevention of DNA replication stress during general DNA replication and in replication-related G2/M-checkpoint regulation. In addition to demonstrating the pathogenicity of identified SPRTN mutations, our findings provide a molecular explanation of how SPRTN dysfunction causes accelerated aging and susceptibility toward carcinoma.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




Comment in
-
SPRTN is a new player in an old story.Nat Genet. 2014 Nov;46(11):1155-7. doi: 10.1038/ng.3125. Nat Genet. 2014. PMID: 25352099
References
-
- Davis EJ, et al. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat Struct Mol Biol. 2012;19:1093–1100. - PubMed
-
- Mosbech A, et al. DVC1 (C1orf124) is a DNA damage–targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat Struct Mol Biol. 2012;19:1084–1092. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- UniProtKB/Q9H040
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

