DataSHIELD: taking the analysis to the data, not the data to the analysis
- PMID: 25261970
- PMCID: PMC4276062
- DOI: 10.1093/ije/dyu188
DataSHIELD: taking the analysis to the data, not the data to the analysis
Abstract
Background: Research in modern biomedicine and social science requires sample sizes so large that they can often only be achieved through a pooled co-analysis of data from several studies. But the pooling of information from individuals in a central database that may be queried by researchers raises important ethico-legal questions and can be controversial. In the UK this has been highlighted by recent debate and controversy relating to the UK's proposed 'care.data' initiative, and these issues reflect important societal and professional concerns about privacy, confidentiality and intellectual property. DataSHIELD provides a novel technological solution that can circumvent some of the most basic challenges in facilitating the access of researchers and other healthcare professionals to individual-level data.
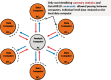
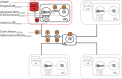
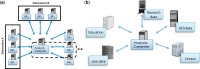
Methods: Commands are sent from a central analysis computer (AC) to several data computers (DCs) storing the data to be co-analysed. The data sets are analysed simultaneously but in parallel. The separate parallelized analyses are linked by non-disclosive summary statistics and commands transmitted back and forth between the DCs and the AC. This paper describes the technical implementation of DataSHIELD using a modified R statistical environment linked to an Opal database deployed behind the computer firewall of each DC. Analysis is controlled through a standard R environment at the AC.
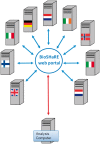
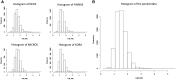
Results: Based on this Opal/R implementation, DataSHIELD is currently used by the Healthy Obese Project and the Environmental Core Project (BioSHaRE-EU) for the federated analysis of 10 data sets across eight European countries, and this illustrates the opportunities and challenges presented by the DataSHIELD approach.
Conclusions: DataSHIELD facilitates important research in settings where: (i) a co-analysis of individual-level data from several studies is scientifically necessary but governance restrictions prohibit the release or sharing of some of the required data, and/or render data access unacceptably slow; (ii) a research group (e.g. in a developing nation) is particularly vulnerable to loss of intellectual property-the researchers want to fully share the information held in their data with national and international collaborators, but do not wish to hand over the physical data themselves; and (iii) a data set is to be included in an individual-level co-analysis but the physical size of the data precludes direct transfer to a new site for analysis.
Keywords: DataSHIELD; ELSI; bioinformatics; confidentiality; disclosure; distributed computing; intellectual property; pooled analysis; privacy.
© The Author 2014; all rights reserved. Published by Oxford University Press on behalf of the International Epidemiological Association.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

