Progranulin protects against amyloid β deposition and toxicity in Alzheimer's disease mouse models
- PMID: 25261995
- PMCID: PMC4196723
- DOI: 10.1038/nm.3672
Progranulin protects against amyloid β deposition and toxicity in Alzheimer's disease mouse models
Abstract
Haploinsufficiency of the progranulin (PGRN) gene (GRN) causes familial frontotemporal lobar degeneration (FTLD) and modulates an innate immune response in humans and in mouse models. GRN polymorphism may be linked to late-onset Alzheimer's disease (AD). However, the role of PGRN in AD pathogenesis is unknown. Here we show that PGRN inhibits amyloid β (Aβ) deposition. Selectively reducing microglial expression of PGRN in AD mouse models impaired phagocytosis, increased plaque load threefold and exacerbated cognitive deficits. Lentivirus-mediated PGRN overexpression lowered plaque load in AD mice with aggressive amyloid plaque pathology. Aβ plaque load correlated negatively with levels of hippocampal PGRN, showing the dose-dependent inhibitory effects of PGRN on plaque deposition. PGRN also protected against Aβ toxicity. Lentivirus-mediated PGRN overexpression prevented spatial memory deficits and hippocampal neuronal loss in AD mice. The protective effects of PGRN against Aβ deposition and toxicity have important therapeutic implications. We propose enhancing PGRN as a potential treatment for PGRN-deficient FTLD and AD.
Figures
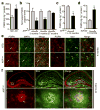


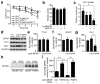

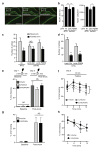
Comment in
-
Understanding the role of progranulin in Alzheimer's disease.Nat Med. 2014 Oct;20(10):1099-100. doi: 10.1038/nm.3712. Nat Med. 2014. PMID: 25295938 No abstract available.
References
-
- Daniel R, He Z, Carmichael KP, Halper J, Bateman A. Cellular localization of gene expression for progranulin. J Histochem Cytochem. 2000;48:999–1009. - PubMed
-
- Daniel R, Daniels E, He Z, Bateman A. Progranulin (acrogranin/PC cell-derived growth factor/granulin-epithelin precursor) is expressed in the placenta, epidermis, microvasculature, and brain during murine development. Dev Dyn. 2003;227:593–599. - PubMed
-
- Petkau TL, et al. Progranulin expression in the developing and adult murine brain. J Comp Neurol. 2010;518:3931–3947. - PubMed
-
- Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. - PubMed
-
- Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

