Modulation of oligodendrocyte generation during a critical temporal window after NG2 cell division
- PMID: 25262495
- PMCID: PMC4275302
- DOI: 10.1038/nn.3815
Modulation of oligodendrocyte generation during a critical temporal window after NG2 cell division
Abstract
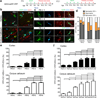
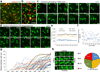
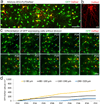
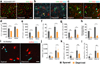
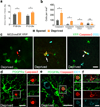
Oligodendrocytes in the mammalian brain are continuously generated from NG2 cells throughout postnatal life. However, it is unclear when the decision is made for NG2 cells to self-renew or differentiate into oligodendrocytes after cell division. Using a combination of in vivo and ex vivo imaging and fate analysis of proliferated NG2 cells in fixed tissue, we demonstrate that in the postnatal developing mouse brain, the majority of divided NG2 cells differentiate into oligodendrocytes during a critical age-specific temporal window of 3-8 d. Notably, within this time period, damage to myelin and oligodendrocytes accelerated oligodendrocyte differentiation from divided cells, and whisker removal decreased the survival of divided cells in the deprived somatosensory cortex. These findings indicate that during the critical temporal window of plasticity, the fate of divided NG2 cells is sensitive to modulation by external signals.
Figures


References
-
- Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat. Rev. Neurosci. 2009;10:9–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

