Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants
- PMID: 25262649
- PMCID: PMC4185121
- DOI: 10.1016/j.ajhg.2014.09.001
Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants
Abstract
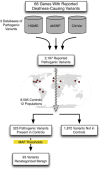
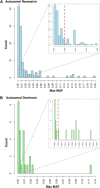
Ethnic-specific differences in minor allele frequency impact variant categorization for genetic screening of nonsyndromic hearing loss (NSHL) and other genetic disorders. We sought to evaluate all previously reported pathogenic NSHL variants in the context of a large number of controls from ethnically distinct populations sequenced with orthogonal massively parallel sequencing methods. We used HGMD, ClinVar, and dbSNP to generate a comprehensive list of reported pathogenic NSHL variants and re-evaluated these variants in the context of 8,595 individuals from 12 populations and 6 ethnically distinct major human evolutionary phylogenetic groups from three sources (Exome Variant Server, 1000 Genomes project, and a control set of individuals created for this study, the OtoDB). Of the 2,197 reported pathogenic deafness variants, 325 (14.8%) were present in at least one of the 8,595 controls, indicating a minor allele frequency (MAF) > 0.00006. MAFs ranged as high as 0.72, a level incompatible with pathogenicity for a fully penetrant disease like NSHL. Based on these data, we established MAF thresholds of 0.005 for autosomal-recessive variants (excluding specific variants in GJB2) and 0.0005 for autosomal-dominant variants. Using these thresholds, we recategorized 93 (4.2%) of reported pathogenic variants as benign. Our data show that evaluation of reported pathogenic deafness variants using variant MAFs from multiple distinct ethnicities and sequenced by orthogonal methods provides a powerful filter for determining pathogenicity. The proposed MAF thresholds will facilitate clinical interpretation of variants identified in genetic testing for NSHL. All data are publicly available to facilitate interpretation of genetic variants causing deafness.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
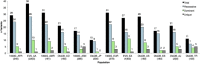
References
-
- Walsh T., Lee M.K., Casadei S., Thornton A.M., Stray S.M., Pennil C., Nord A.S., Mandell J.B., Swisher E.M., King M.C. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2010;107:12629–12633. - PMC - PubMed
-
- Shanks M.E., Downes S.M., Copley R.R., Lise S., Broxholme J., Hudspith K.A., Kwasniewska A., Davies W.I., Hankins M.W., Packham E.R. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur. J. Hum. Genet. 2012;21:274–280. - PMC - PubMed
-
- Smith R.J., Bale J.F., Jr., White K.R. Sensorineural hearing loss in children. Lancet. 2005;365:879–890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

