De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies
- PMID: 25262651
- PMCID: PMC4185114
- DOI: 10.1016/j.ajhg.2014.08.013
De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies
Erratum in
-
De Novo Mutations in Synaptic Transmission Genes Including DNM1 Cause Epileptic Encephalopathies.Am J Hum Genet. 2017 Jan 5;100(1):179. doi: 10.1016/j.ajhg.2016.12.012. Am J Hum Genet. 2017. PMID: 28061363 Free PMC article. No abstract available.
Abstract
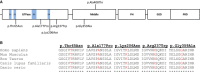
Emerging evidence indicates that epileptic encephalopathies are genetically highly heterogeneous, underscoring the need for large cohorts of well-characterized individuals to further define the genetic landscape. Through a collaboration between two consortia (EuroEPINOMICS and Epi4K/EPGP), we analyzed exome-sequencing data of 356 trios with the "classical" epileptic encephalopathies, infantile spasms and Lennox Gastaut syndrome, including 264 trios previously analyzed by the Epi4K/EPGP consortium. In this expanded cohort, we find 429 de novo mutations, including de novo mutations in DNM1 in five individuals and de novo mutations in GABBR2, FASN, and RYR3 in two individuals each. Unlike previous studies, this cohort is sufficiently large to show a significant excess of de novo mutations in epileptic encephalopathy probands compared to the general population using a likelihood analysis (p = 8.2 × 10(-4)), supporting a prominent role for de novo mutations in epileptic encephalopathies. We bring statistical evidence that mutations in DNM1 cause epileptic encephalopathy, find suggestive evidence for a role of three additional genes, and show that at least 12% of analyzed individuals have an identifiable causal de novo mutation. Strikingly, 75% of mutations in these probands are predicted to disrupt a protein involved in regulating synaptic transmission, and there is a significant enrichment of de novo mutations in genes in this pathway in the entire cohort as well. These findings emphasize an important role for synaptic dysregulation in epileptic encephalopathies, above and beyond that caused by ion channel dysfunction.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Berg A.T., Berkovic S.F., Brodie M.J., Buchhalter J., Cross J.H., van Emde Boas W., Engel J., French J., Glauser T.A., Mathern G.W. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005-2009. Epilepsia. 2010;51:676–685. - PubMed
-
- Kodera H., Nakamura K., Osaka H., Maegaki Y., Haginoya K., Mizumoto S., Kato M., Okamoto N., Iai M., Kondo Y. De novo mutations in SLC35A2 encoding a UDP-galactose transporter cause early-onset epileptic encephalopathy. Hum. Mutat. 2013;34:1708–1714. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

