Genetic alterations of protein tyrosine phosphatases in human cancers
- PMID: 25263441
- PMCID: PMC4377308
- DOI: 10.1038/onc.2014.326
Genetic alterations of protein tyrosine phosphatases in human cancers
Abstract
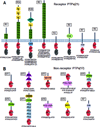
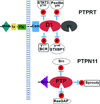
Protein tyrosine phosphatases (PTPs) are enzymes that remove phosphate from tyrosine residues in proteins. Recent whole-exome sequencing of human cancer genomes reveals that many PTPs are frequently mutated in a variety of cancers. Among these mutated PTPs, PTP receptor T (PTPRT) appears to be the most frequently mutated PTP in human cancers. Beside PTPN11, which functions as an oncogene in leukemia, genetic and functional studies indicate that most of mutant PTPs are tumor suppressor genes. Identification of the substrates and corresponding kinases of the mutant PTPs may provide novel therapeutic targets for cancers harboring these mutant PTPs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Julien SG, Dubé N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat Rev Cancer. 2011;11:35–49. - PubMed
-
- Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. - PubMed
-
- Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

