Evolutions in fragment-based drug design: the deconstruction-reconstruction approach
- PMID: 25263697
- PMCID: PMC4305461
- DOI: 10.1016/j.drudis.2014.09.015
Evolutions in fragment-based drug design: the deconstruction-reconstruction approach
Abstract
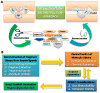
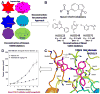
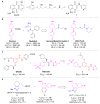
Recent advances in the understanding of molecular recognition and protein-ligand interactions have facilitated rapid development of potent and selective ligands for therapeutically relevant targets. Over the past two decades, a variety of useful approaches and emerging techniques have been developed to promote the identification and optimization of leads that have high potential for generating new therapeutic agents. Intriguingly, the innovation of a fragment-based drug design (FBDD) approach has enabled rapid and efficient progress in drug discovery. In this critical review, we focus on the construction of fragment libraries and the advantages and disadvantages of various fragment-based screening (FBS) for constructing such libraries. We also highlight the deconstruction-reconstruction strategy by utilizing privileged fragments of reported ligands.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures

References
-
- Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discov. 2007;6:211–219. - PubMed
-
- Fattori D, et al. Fragment-based approach to drug lead discovery: overview and advances in various techniques. Drugs R D. 2008;9:217–227. - PubMed
-
- Erlanson DA. Introduction to fragment-based drug discovery. Top Curr Chem. 2012;317:1–32. - PubMed
-
- Hajduk PJ. Puzzling through fragment-based drug design. Nat Chem Biol. 2006;2:658–659. - PubMed
-
- Hajduk PJ. Fragment-based drug design: how big is too big? J Med Chem. 2006;49:6972–6976. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

