Identification of differentially expressed genes in breast muscle and skin fat of postnatal Pekin duck
- PMID: 25264787
- PMCID: PMC4180276
- DOI: 10.1371/journal.pone.0107574
Identification of differentially expressed genes in breast muscle and skin fat of postnatal Pekin duck
Abstract
Lean-type Pekin duck is a commercial breed that has been obtained through long-term selection. Investigation of the differentially expressed genes in breast muscle and skin fat at different developmental stages will contribute to a comprehensive understanding of the potential mechanisms underlying the lean-type Pekin duck phenotype. In the present study, RNA-seq was performed on breast muscle and skin fat at 2-, 4- and 6-weeks of age. More than 89% of the annotated duck genes were covered by our RNA-seq dataset. Thousands of differentially expressed genes, including many important genes involved in the regulation of muscle development and fat deposition, were detected through comparison of the expression levels in the muscle and skin fat of the same time point, or the same tissue at different time points. KEGG pathway analysis showed that the differentially expressed genes clustered significantly in many muscle development and fat deposition related pathways such as MAPK signaling pathway, PPAR signaling pathway, Calcium signaling pathway, Fat digestion and absorption, and TGF-beta signaling pathway. The results presented here could provide a basis for further investigation of the mechanisms involved in muscle development and fat deposition in Pekin duck.
Conflict of interest statement
Figures
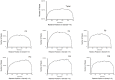

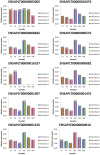

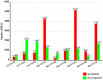
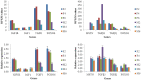
Similar articles
-
RNA-Seq transcriptome analysis of breast muscle in Pekin ducks supplemented with the dietary probiotic Clostridium butyricum.BMC Genomics. 2018 Nov 28;19(1):844. doi: 10.1186/s12864-018-5261-1. BMC Genomics. 2018. PMID: 30486769 Free PMC article.
-
Gene expression profiling in Pekin duck embryonic breast muscle.PLoS One. 2017 May 4;12(5):e0174612. doi: 10.1371/journal.pone.0174612. eCollection 2017. PLoS One. 2017. PMID: 28472139 Free PMC article.
-
Characterization and comparative transcriptomic analysis of skeletal muscle in female Pekin duck and Hanzhong Ma duck during different growth stages using RNA-seq.Poult Sci. 2023 Dec;102(12):103122. doi: 10.1016/j.psj.2023.103122. Epub 2023 Sep 18. Poult Sci. 2023. PMID: 37832186 Free PMC article.
-
Integrating genome and transcriptome profiling for elucidating the mechanism of muscle growth and lipid deposition in Pekin ducks.Sci Rep. 2017 Jun 19;7(1):3837. doi: 10.1038/s41598-017-04178-7. Sci Rep. 2017. PMID: 28630415 Free PMC article.
-
Skeletal Muscle Transcriptome Analysis of Hanzhong Ma Duck at Different Growth Stages Using RNA-Seq.Biomolecules. 2021 Feb 19;11(2):315. doi: 10.3390/biom11020315. Biomolecules. 2021. PMID: 33669581 Free PMC article.
Cited by
-
RNA-Seq transcriptome analysis of breast muscle in Pekin ducks supplemented with the dietary probiotic Clostridium butyricum.BMC Genomics. 2018 Nov 28;19(1):844. doi: 10.1186/s12864-018-5261-1. BMC Genomics. 2018. PMID: 30486769 Free PMC article.
-
Transcriptional Insights into Key Genes and Pathways Underlying Muscovy Duck Subcutaneous Fat Deposition at Different Developmental Stages.Animals (Basel). 2021 Jul 15;11(7):2099. doi: 10.3390/ani11072099. Animals (Basel). 2021. PMID: 34359227 Free PMC article.
-
Gene expression profiling in Pekin duck embryonic breast muscle.PLoS One. 2017 May 4;12(5):e0174612. doi: 10.1371/journal.pone.0174612. eCollection 2017. PLoS One. 2017. PMID: 28472139 Free PMC article.
-
Analysis of Anasplatyrhynchos genome resequencing data reveals genetic signatures of artificial selection.PLoS One. 2019 Feb 8;14(2):e0211908. doi: 10.1371/journal.pone.0211908. eCollection 2019. PLoS One. 2019. PMID: 30735526 Free PMC article.
-
Nutritional requirements of meat-type and egg-type ducks: what do we know?J Anim Sci Biotechnol. 2018 Jan 16;9:1. doi: 10.1186/s40104-017-0217-x. eCollection 2018. J Anim Sci Biotechnol. 2018. PMID: 29372052 Free PMC article. Review.
References
-
- Xu TS, Huang W, Zhang XH, Ye BG, Zhou HL, et al. (2012) Identification and characterization of genes related to the development of breast muscles in Pekin duck. Mol Biol Rep 39: 7647–7655. - PubMed
-
- Smith JH (1963) Relation of body size to muscle cell size and number in the chicken. Poult Sci 42: 283–290.
-
- Fowler SP, Campion DR, Marks HL, Reagan JO (1980) An analysis of skeletal-muscle response to selection for rapid growth in japanese quail (Coturnix-Coturnix-Japonica). Growth 44: 235–252. - PubMed
-
- Braun T, Gautel M (2011) Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Bio 12: 349–361. - PubMed
-
- Chakravarthy MV, Fiorotto ML, Schwartz RJ, Booth FW (2001) Long-term insulin-like growth factor-I expression in skeletal muscles attenuates the enhanced in vitro proliferation ability of the resident satellite cells in transgenic mice. Mech Ageing Dev 122: 1303–1320. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

