Hybrid Bayesian-rank integration approach improves the predictive power of genomic dataset aggregation
- PMID: 25266226
- PMCID: PMC4287939
- DOI: 10.1093/bioinformatics/btu518
Hybrid Bayesian-rank integration approach improves the predictive power of genomic dataset aggregation
Abstract
Motivation: Modern molecular technologies allow the collection of large amounts of high-throughput data on the functional attributes of genes. Often multiple technologies and study designs are used to address the same biological question such as which genes are overexpressed in a specific disease state. Consequently, there is considerable interest in methods that can integrate across datasets to present a unified set of predictions.
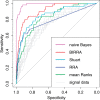
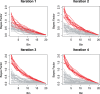
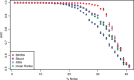
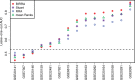
Results: An important aspect of data integration is being able to account for the fact that datasets may differ in how accurately they capture the biological signal of interest. While many methods to address this problem exist, they always rely either on dataset internal statistics, which reflect data structure and not necessarily biological relevance, or external gold standards, which may not always be available. We present a new rank aggregation method for data integration that requires neither external standards nor internal statistics but relies on Bayesian reasoning to assess dataset relevance. We demonstrate that our method outperforms established techniques and significantly improves the predictive power of rank-based aggregations. We show that our method, which does not require an external gold standard, provides reliable estimates of dataset relevance and allows the same set of data to be integrated differently depending on the specific signal of interest.
Availability: The method is implemented in R and is freely available at http://www.pitt.edu/~mchikina/BIRRA/.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Akey JM, et al. On the design and analysis of gene expression studies in human populations. Nat. Genet. 2007;39:807–808. ; author reply 808–809. - PubMed
-
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell. 2004;15:57–67. - PubMed
-
- Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

