U2AF1 mutations alter splice site recognition in hematological malignancies
- PMID: 25267526
- PMCID: PMC4317169
- DOI: 10.1101/gr.181016.114
U2AF1 mutations alter splice site recognition in hematological malignancies
Abstract
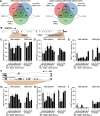
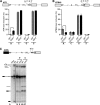
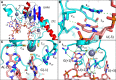
Whole-exome sequencing studies have identified common mutations affecting genes encoding components of the RNA splicing machinery in hematological malignancies. Here, we sought to determine how mutations affecting the 3' splice site recognition factor U2AF1 alter its normal role in RNA splicing. We find that U2AF1 mutations influence the similarity of splicing programs in leukemias, but do not give rise to widespread splicing failure. U2AF1 mutations cause differential splicing of hundreds of genes, affecting biological pathways such as DNA methylation (DNMT3B), X chromosome inactivation (H2AFY), the DNA damage response (ATR, FANCA), and apoptosis (CASP8). We show that U2AF1 mutations alter the preferred 3' splice site motif in patients, in cell culture, and in vitro. Mutations affecting the first and second zinc fingers give rise to different alterations in splice site preference and largely distinct downstream splicing programs. These allele-specific effects are consistent with a computationally predicted model of U2AF1 in complex with RNA. Our findings suggest that U2AF1 mutations contribute to pathogenesis by causing quantitative changes in splicing that affect diverse cellular pathways, and give insight into the normal function of U2AF1's zinc finger domains.
© 2015 Ilagan et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, et al. . 2014. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS ONE 9: e87361. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- P30 CA015704/CA/NCI NIH HHS/United States
- NIH/NCI P30 CA015704/CA/NCI NIH HHS/United States
- P30 DK056465/DK/NIDDK NIH HHS/United States
- T32 CA009657/CA/NCI NIH HHS/United States
- R01 GM088277/GM/NIGMS NIH HHS/United States
- NIH/NHLBI U01 HL099993/PHS HHS/United States
- NIH/NIDDK P30 DK056465/PHS HHS/United States
- U01 HL099993/HL/NHLBI NIH HHS/United States
- T32CA009657/CA/NCI NIH HHS/United States
- NIH/NIDDK K08 DK082783/PHS HHS/United States
- NIH/NIGMS R01 GM088277/PHS HHS/United States
- K08 DK082783/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
