Multifunctional RNA nanoparticles
- PMID: 25267559
- PMCID: PMC4189619
- DOI: 10.1021/nl502385k
Multifunctional RNA nanoparticles
Abstract
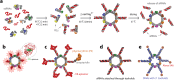
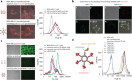
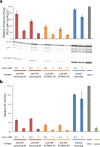
Our recent advancements in RNA nanotechnology introduced novel nanoscaffolds (nanorings); however, the potential of their use for biomedical applications was never fully revealed. As presented here, besides functionalization with multiple different short interfering RNAs for combinatorial RNA interference (e.g., against multiple HIV-1 genes), nanorings also allow simultaneous embedment of assorted RNA aptamers, fluorescent dyes, proteins, as well as recently developed RNA-DNA hybrids aimed to conditionally activate multiple split functionalities inside cells.
Keywords: RNA interference; RNA nanoparticles; RNA nanotechnology; RNA−DNA hybrid reassociation; aptamers.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

