Structural basis of cellular dNTP regulation by SAMHD1
- PMID: 25267621
- PMCID: PMC4205617
- DOI: 10.1073/pnas.1412289111
Structural basis of cellular dNTP regulation by SAMHD1
Abstract
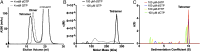
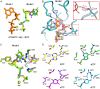
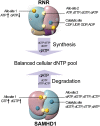
The sterile alpha motif and HD domain-containing protein 1 (SAMHD1), a dNTPase, prevents the infection of nondividing cells by retroviruses, including HIV, by depleting the cellular dNTP pool available for viral reverse transcription. SAMHD1 is a major regulator of cellular dNTP levels in mammalian cells. Mutations in SAMHD1 are associated with chronic lymphocytic leukemia (CLL) and the autoimmune condition Aicardi Goutières syndrome (AGS). The dNTPase activity of SAMHD1 can be regulated by dGTP, with which SAMHD1 assembles into catalytically active tetramers. Here we present extensive biochemical and structural data that reveal an exquisite activation mechanism of SAMHD1 via combined action of both GTP and dNTPs. We obtained 26 crystal structures of SAMHD1 in complex with different combinations of GTP and dNTP mixtures, which depict the full spectrum of GTP/dNTP binding at the eight allosteric and four catalytic sites of the SAMHD1 tetramer. Our data demonstrate how SAMHD1 is activated by binding of GTP or dGTP at allosteric site 1 and a dNTP of any type at allosteric site 2. Our enzymatic assays further reveal a robust regulatory mechanism of SAMHD1 activity, which bares resemblance to that of the ribonuclease reductase responsible for cellular dNTP production. These results establish a complete framework for a mechanistic understanding of the important functions of SAMHD1 in the regulation of cellular dNTP levels, as well as in HIV restriction and the pathogenesis of CLL and AGS.
Keywords: HIV restriction factor; allosteric regulation; dNTP metabolism; tetramerization; triphosphohydrolase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

