Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins
- PMID: 25267629
- PMCID: PMC4205638
- DOI: 10.1073/pnas.1417138111
Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins
Abstract
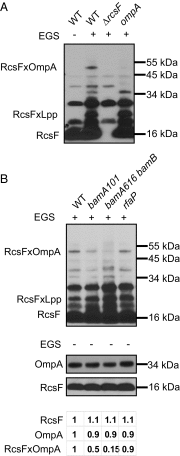
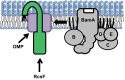
RcsF (regulator of capsule synthesis) is an outer membrane (OM) lipoprotein that functions to sense defects such as changes in LPS. However, LPS is found in the outer leaflet, and RcsF was thought to be tethered to the inner leaflet by its lipidated N terminus, raising the question of how it monitors LPS. We show that RcsF has a transmembrane topology with the lipidated N terminus on the cell surface and the C-terminal signaling domain in the periplasm. Strikingly, the short, unstructured, charged transmembrane domain is threaded through the lumen of β-barrel OM proteins where it is protected from the hydrophobic membrane interior. We present evidence that these unusual complexes, which contain one protein inside another, are formed by the Bam complex that assembles all β-barrel proteins in the OM. The ability of the Bam complex to expose lipoproteins at the cell surface underscores the mechanistic versatility of the β-barrel assembly machine.
Keywords: envelope stress response; membrane biogenesis; protein folding; signal transduction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

