Erosion of functional independence early in the evolution of a microbial mutualism
- PMID: 25267659
- PMCID: PMC4205623
- DOI: 10.1073/pnas.1407986111
Erosion of functional independence early in the evolution of a microbial mutualism
Abstract
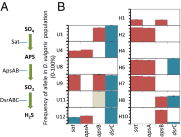
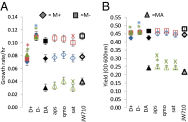
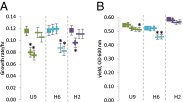
Many species have evolved to function as specialized mutualists, often to the detriment of their ability to survive independently. However, there are few, if any, well-controlled observations of the evolutionary processes underlying the genesis of new mutualisms. Here, we show that within the first 1,000 generations of initiating independent syntrophic interactions between a sulfate reducer (Desulfovibrio vulgaris) and a hydrogenotrophic methanogen (Methanococcus maripaludis), D. vulgaris frequently lost the capacity to grow by sulfate respiration, thus losing the primary physiological attribute of the genus. The loss of sulfate respiration was a consequence of mutations in one or more of three key genes in the pathway for sulfate respiration, required for sulfate activation (sat) and sulfate reduction to sulfite (apsA or apsB). Because loss-of-function mutations arose rapidly and independently in replicated experiments, and because these mutations were correlated with enhanced growth rate and productivity, gene loss could be attributed to natural selection, even though these mutations should significantly restrict the independence of the evolved D. vulgaris. Together, these data present an empirical demonstration that specialization for a mutualistic interaction can evolve by natural selection shortly after its origin. They also demonstrate that a sulfate-reducing bacterium can readily evolve to become a specialized syntroph, a situation that may have often occurred in nature.
Keywords: coevolution; experimental evolution; sulfate-reducing prokaryote; syntrophy; trade-offs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Pellmyr O, Thompson JN, Brown JM, Harrison RG. Evolution of pollination and mutualism in the yucca moth lineage. Am Nat. 1996;148(5):827–847.
-
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. - PubMed
-
- Sachs JL, Simms EL. Pathways to mutualism breakdown. Trends Ecol Evol. 2006;21(10):585–592. - PubMed
-
- Losos JB. Seeing the forest for the trees: The limitations of phylogenies in comparative biology. (American Society of Naturalists Address) Am Nat. 2011;177(6):709–727. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

