Potency and resistance analysis of hepatitis C virus NS5B polymerase inhibitor BMS-791325 on all major genotypes
- PMID: 25267677
- PMCID: PMC4249524
- DOI: 10.1128/AAC.03851-14
Potency and resistance analysis of hepatitis C virus NS5B polymerase inhibitor BMS-791325 on all major genotypes
Abstract
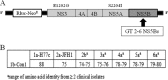
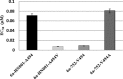
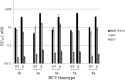
BMS-791325 is a hepatitis C virus (HCV) inhibitor binding to the thumb domain of the NS5B RNA-dependent RNA polymerase. BMS-791325 is well characterized in genotype 1 (GT1) and exhibits good inhibitory activity (50% effective concentration [EC50], <10 nM) against hybrid replicons containing patient NS5B sequences from GT3a, -4a, and -5a while potency against GT2 is significantly reduced (J. A. Lemm et al., Antimicrob. Agents Chemother. 58:3485-3495, 2014, doi:http://dx.doi.org/10.1128/AAC.02495-13). BMS-791325 potency against GT6a hybrid replicons is more variable, with two of three hybrid clones having EC50s similar to that for GT1 while a third patient clone was ∼ 10 times less susceptible to BMS-791325. To characterize the resistance profile of BMS-791325 beyond GT1, curing studies were performed across GT1a and -3a to -6a and demonstrated that GT1a has the highest resistance barrier versus BMS-791325 while GT6a has the lowest. Selection of GT3 to -6 NS5B chimeric replicon cells at different concentrations of BMS-791325 revealed substitutions in the thumb domain of NS5B at residues 494 and 495 that conferred different levels of resistance to BMS-791325 but remained susceptible to NS5A or NS3 protease inhibitors. In addition, we demonstrate that the reduced potency of BMS-791325 against one GT6a patient is due to an A494 polymorphism present in ∼ 21% of sequences in the European HCV database. The results from this report suggest that BMS-791325 is a candidate for combination treatment of HCV GT3 to -6 chronic infections, and the resistance profiles identified will provide useful information for future clinical development.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

