The genome landscape of the african green monkey kidney-derived vero cell line
- PMID: 25267831
- PMCID: PMC4263300
- DOI: 10.1093/dnares/dsu029
The genome landscape of the african green monkey kidney-derived vero cell line
Abstract
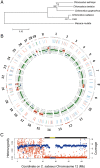
Continuous cell lines that originate from mammalian tissues serve as not only invaluable tools for life sciences, but also important animal cell substrates for the production of various types of biological pharmaceuticals. Vero cells are susceptible to various types of microbes and toxins and have widely contributed to not only microbiology, but also the production of vaccines for human use. We here showed the genome landscape of a Vero cell line, in which 25,877 putative protein-coding genes were identified in the 2.97-Gb genome sequence. A homozygous ∼9-Mb deletion on chromosome 12 caused the loss of the type I interferon gene cluster and cyclin-dependent kinase inhibitor genes in Vero cells. In addition, an ∼59-Mb loss of heterozygosity around this deleted region suggested that the homozygosity of the deletion was established by a large-scale conversion. Moreover, a genomic analysis of Vero cells revealed a female Chlorocebus sabaeus origin and proviral variations of the endogenous simian type D retrovirus. These results revealed the genomic basis for the non-tumourigenic permanent Vero cell lineage susceptible to various pathogens and will be useful for generating new sub-lines and developing new tools in the quality control of Vero cells.
Keywords: Vero cell; animal cell substrate; infectious diseases; vaccine; whole genome.
© The Author 2014. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures



References
-
- Yasumura Y., Kawakita Y. Studies on SV40 in tissue culture: preliminary step for cancer reserach in vitro (in Japanese) Nihon Rinsho. 1963;21:1201–15.
-
- Yasumura Y., Kawakita Y. Studies on SV40 in tissue culture: preliminary step for cancer research in vitro. In: Simizu, B., Terasima T., editors. VERO cells: origin, properties and biomedical applications. Chiba, Japan: Department of Microbiology School of Medicine Chiba University; 1988. pp. 1–19.
-
- Sasaki K., Makino S., Kasahara S. Studies on measles virus. II. Propagation in two established simian renal cell lines and development of a plaque assay. Kitasato Arch. Exp. Med. 1964;37:27–42. - PubMed
-
- Rhim J.S., Schell K. Cytopathic and plaque assay of rubella virus in a line of African green monkey kidney cells (Vero) Proc. Soc. Exp. Biol. Med. 1967;125:602–6. - PubMed
-
- Liebhaber H., Riordan J.T., Horstmann D.M. Replication of rubella virus in a continuous line of African green monkey kidney cells (Vero) Proc. Soc. Exp. Biol. Med. 1967;125:636–43. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

