Covalent small molecule inhibitors of Ca(2+)-bound S100B
- PMID: 25268459
- PMCID: PMC4211652
- DOI: 10.1021/bi5005552
Covalent small molecule inhibitors of Ca(2+)-bound S100B
Abstract
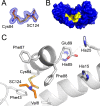
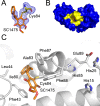
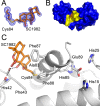
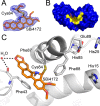
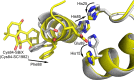
Elevated levels of the tumor marker S100B are observed in malignant melanoma, and this EF-hand-containing protein was shown to directly bind wild-type (wt) p53 in a Ca(2+)-dependent manner, dissociate the p53 tetramer, and inhibit its tumor suppression functions. Likewise, inhibiting S100B with small interfering RNA (siRNA(S100B)) is sufficient to restore wild-type p53 levels and its downstream gene products and induce the arrest of cell growth and UV-dependent apoptosis in malignant melanoma. Therefore, it is a goal to develop S100B inhibitors (SBiXs) that inhibit the S100B-p53 complex and restore active p53 in this deadly cancer. Using a structure-activity relationship by nuclear magnetic resonance approach (SAR by NMR), three persistent binding pockets are found on S100B, termed sites 1-3. While inhibitors that simultaneously bind sites 2 and 3 are in place, no molecules that simultaneously bind all three persistent sites are available. For this purpose, Cys84 was used in this study as a potential means to bridge sites 1 and 2 because it is located in a small crevice between these two deeper pockets on the protein. Using a fluorescence polarization competition assay, several Cys84-modified S100B complexes were identified and examined further. For five such SBiX-S100B complexes, crystallographic structures confirmed their covalent binding to Cys84 near site 2 and thus present straightforward chemical biology strategies for bridging sites 1 and 3. Importantly, one such compound, SC1982, showed an S100B-dependent death response in assays with WM115 malignant melanoma cells, so it will be particularly useful for the design of SBiX molecules with improved affinity and specificity.
Figures





References
-
- Kwong L. N.; Davies M. A. (2014) Targeted therapy for melanoma: Rational combinatorial approaches. Oncogene 33, 1–9. - PubMed
-
- Topalian S. L.; Hodi F. S.; Brahmer J. R.; Gettinger S. N.; Smith D. C.; McDermott D. F.; Powderly J. D.; Carvajal R. D.; Sosman J. A.; Atkins M. B.; Leming P. D.; Spigel D. R.; Antonia S. J.; Horn L.; Drake C. G.; Pardoll D. M.; Chen L.; Sharfman W. H.; Anders R. A.; Taube J. M.; McMiller T. L.; Xu H.; Korman A. J.; Jure-Kunkel M.; Agrawal S.; McDonald D.; Kollia G. D.; Gupta A.; Wigginton J. M.; Sznol M. (2012) Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454. - PMC - PubMed
-
- Bollag G.; Hirth P.; Tsai J.; Zhang J.; Ibrahim P. N.; Cho H.; Spevak W.; Zhang C.; Zhang Y.; Habets G.; Burton E. A.; Wong B.; Tsang G.; West B. L.; Powell B.; Shellooe R.; Marimuthu A.; Nguyen H.; Zhang K. Y.; Artis D. R.; Schlessinger J.; Su F.; Higgins B.; Iyer R.; D’Andrea K.; Koehler A.; Stumm M.; Lin P. S.; Lee R. J.; Grippo J.; Puzanov I.; Kim K. B.; Ribas A.; McArthur G. A.; Sosman J. A.; Chapman P. B.; Flaherty K. T.; Xu X.; Nathanson K. L.; Nolop K. (2010) Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature 467, 596–599. - PMC - PubMed
-
- Espinosa E.; Grob J. J.; Dummer R.; Rutkowski P.; Robert C.; Gogas H.; Kefford R.; Eggermont A. M.; Martin Algarra S.; Hauschild A.; Schadendorf D. (2014) Treatment Algorithms in Stage IV Melanoma. American Journal of Therapeutics DOI: 10.1097/MJT.0b013e31829e885c. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

