Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets
- PMID: 25270025
- PMCID: PMC4203973
- DOI: 10.1186/s12915-014-0078-0
Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets
Abstract
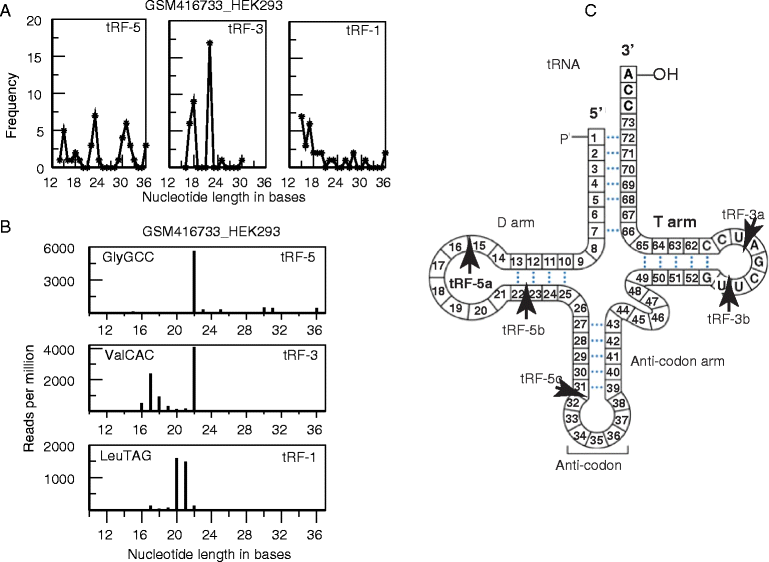
Background: tRFs, 14 to 32 nt long single-stranded RNA derived from mature or precursor tRNAs, are a recently discovered class of small RNA that have been found to be present in diverse organisms at read counts comparable to miRNAs. Currently, there is a debate about their biogenesis and function.
Results: This is the first meta-analysis of tRFs. Analysis of more than 50 short RNA libraries has revealed that tRFs are precisely generated fragments present in all domains of life (bacteria to humans), and are not produced by the miRNA biogenesis pathway. Human PAR-CLIP data shows a striking preference for tRF-5s and tRF-3s to associate with AGO1, 3 and 4 rather than AGO2, and analysis of positional T to C mutational frequency indicates these tRFs associate with Argonautes in a manner similar to miRNAs. The reverse complements of canonical seed positions in these sequences match cross-link centered regions, suggesting these tRF-5s and tRF-3s interact with RNAs in the cell. Consistent with these results, human AGO1 CLASH data contains thousands of tRF-5 and tRF-3 reads chimeric with mRNAs.
Conclusions: tRFs are an abundant class of small RNA present in all domains of life whose biogenesis is distinct from miRNAs. In human HEK293 cells tRFs associate with Argonautes 1, 3 and 4 and not Argonaute 2 which is the main effector protein of miRNA function, but otherwise have very similar properties to miRNAs, indicating tRFs may play a major role in RNA silencing.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

