Pre- and postovulatory aging of murine oocytes affect the transcript level and poly(A) tail length of maternal effect genes
- PMID: 25271735
- PMCID: PMC4182777
- DOI: 10.1371/journal.pone.0108907
Pre- and postovulatory aging of murine oocytes affect the transcript level and poly(A) tail length of maternal effect genes
Abstract
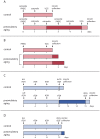
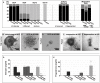
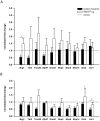
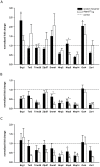
Maternal effect genes code for oocyte proteins that are important for early embryogenesis. Transcription in oocytes does not take place from the onset of meiotic progression until zygotic genome activation. During this period, protein levels are regulated posttranscriptionally, for example by poly(A) tail length. Posttranscriptional regulation may be impaired in preovulatory and postovulatory aged oocytes, caused by delayed ovulation or delayed fertilization, respectively, and may lead to developmental defects. We investigated transcript levels and poly(A) tail length of ten maternal effect genes in in vivo- and in vitro- (follicle culture) grown oocytes after pre- and postovulatory aging. Quantitative RT-PCR was performed using random hexamer-primed cDNA to determine total transcript levels and oligo(dT)16-primed cDNA to analyze poly(A) tail length. Transcript levels of in vivo preovulatory-aged oocytes remained stable except for decreases in Brg1 and Tet3. Most genes investigated showed a tendency towards increased poly(A) content. Polyadenylation of in vitro preovulatory-aged oocytes was also increased, along with transcript level declines of Trim28, Nlrp2, Nlrp14 and Zar1. In contrast to preovulatory aging, postovulatory aging of in vivo- and in vitro-grown oocytes led to a shortening of poly(A) tails. Postovulatory aging of in vivo-grown oocytes resulted in deadenylation of Nlrp5 after 12 h, and deadenylation of 4 further genes (Tet3, Trim28, Dnmt1, Oct4) after 24 h. Similarly, transcripts of in vitro-grown oocytes were deadenylated after 12 h of postovulatory aging (Tet3, Trim28, Zfp57, Dnmt1, Nlrp5, Zar1). This impact of aging on poly(A) tail length may affect the timed translation of maternal effect gene transcripts and thereby contribute to developmental defects.
Conflict of interest statement
Figures
References
-
- Kang MK, Han SJ (2011) Post-transcriptional and post-translational regulation during mouse oocyte maturation. BMB Rep 44: 147–157. - PubMed
-
- Clarke HJ (2012) Post-transcriptional control of gene expression during mouse oogenesis. Berlin Heidelberg: Springer-Verlag. - PubMed
-
- Weill L, Belloc E, Bava FA, Mendez R (2012) Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol 19: 577–585. - PubMed
-
- McGrew LL, Dworkin-Rastl E, Dworkin MB, Richter JD (1989) Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev 3: 803–815. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

