Engineering an enhanced, thermostable, monomeric bacterial luciferase gene as a reporter in plant protoplasts
- PMID: 25271765
- PMCID: PMC4182741
- DOI: 10.1371/journal.pone.0107885
Engineering an enhanced, thermostable, monomeric bacterial luciferase gene as a reporter in plant protoplasts
Abstract
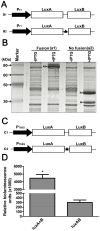
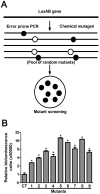
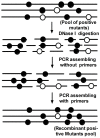
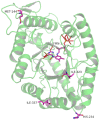
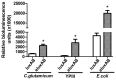
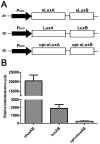
The application of the luxCDABE operon of the bioluminescent bacterium Photorhabdus luminescens as a reporter has been published for bacteria, yeast and mammalian cells. We report here the optimization of fused luxAB (the bacterial luciferase heterodimeric enzyme) expression, quantum yield and its application as a reporter gene in plant protoplasts. The fused luxAB gene was mutated by error prone PCR or chemical mutagenesis and screened for enhanced luciferase activity utilizing decanal as substrate. Positive luxAB mutants with superior quantum yield were subsequently shuffled by DNase I digestion and PCR assembly for generation of recombinants with additional increases in luciferase activity in bacteria. The coding sequence of the best recombinant, called eluxAB, was then optimized further to conform to Arabidopsis (Arabidopsis thaliana) codon usage. A plant expression vector of the final, optimized eluxAB gene (opt-eluxAB) was constructed and transformed into protoplasts of Arabidopsis and maize (Zea mays). Luciferase activity was dramatically increased for opt-eluxAB compared to the original luxAB in Arabidopsis and maize cells. The opt-eluxAB driven by two copies of the 35S promoter expresses significantly higher than that driven by a single copy. These results indicate that the eluxAB gene can be used as a reporter in plant protoplasts. To our knowledge, this is the first report to engineer the bacterium Photorhabdus luminescens luciferase luxAB as a reporter by directed evolution which paved the way for further improving the luxAB reporter in the future.
Conflict of interest statement
Figures


References
-
- Xiong AS, Peng RH, Zhuang J, Davies J, Zhang J, et al. (2012) Advances in directed molecular evolution of reporter genes. Crit Rev Biotechnol 32: 133–142. - PubMed
-
- DiGrazia PM, King JM, Blackburn JW, Applegate BA, Bienkowski PR, et al. (1991) Dynamic response of naphthalene biodegradation in a continuous flow soil slurry reactor. Biodegradation 2: 81–91. - PubMed
-
- Monshausen GB (2012) Visualizing Ca2+ signatures in plants. Curr Opin Plant Biol 15: 677–682. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

