Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection
- PMID: 25274306
- PMCID: PMC4270954
- DOI: 10.1038/nature13818
Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection
Abstract
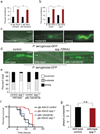
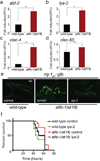
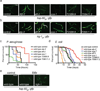
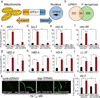
Metazoans identify and eliminate bacterial pathogens in microbe-rich environments such as the intestinal lumen; however, the mechanisms are unclear. Host cells could potentially use intracellular surveillance or stress response programs to detect pathogens that target monitored cellular activities and then initiate innate immune responses. Mitochondrial function is evaluated by monitoring mitochondrial protein import efficiency of the transcription factor ATFS-1, which mediates the mitochondrial unfolded protein response (UPR(mt)). During mitochondrial stress, mitochondrial import is impaired, allowing ATFS-1 to traffic to the nucleus where it mediates a transcriptional response to re-establish mitochondrial homeostasis. Here we examined the role of ATFS-1 in Caenorhabditis elegans during pathogen exposure, because during mitochondrial stress ATFS-1 induced not only mitochondrial protective genes but also innate immune genes that included a secreted lysozyme and anti-microbial peptides. Exposure to the pathogen Pseudomonas aeruginosa caused mitochondrial dysfunction and activation of the UPR(mt). C. elegans lacking atfs-1 were susceptible to P. aeruginosa, whereas hyper-activation of ATFS-1 and the UPR(mt) improved clearance of P. aeruginosa from the intestine and prolonged C. elegans survival in a manner mainly independent of known innate immune pathways. We propose that ATFS-1 import efficiency and the UPR(mt) is a means to detect pathogens that target mitochondria and initiate a protective innate immune response.
Figures
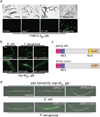
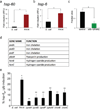
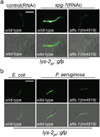
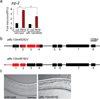


References
-
- Wright G, Terada K, Yano M, Sergeev I, Mori M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp Cell Res. 2001;263:107–117. - PubMed
METHODS REFERENCES
-
- Yoneda T, et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055–4066. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

