Transcriptional regulation by trithorax-group proteins
- PMID: 25274705
- PMCID: PMC4176006
- DOI: 10.1101/cshperspect.a019349
Transcriptional regulation by trithorax-group proteins
Abstract
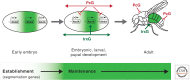
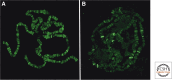
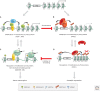
The trithorax group of genes (trxG) was identified in mutational screens that examined developmental phenotypes and suppression of Polycomb mutant phenotypes. The protein products of these genes are primarily involved in gene activation, although some can also have repressive effects. There is no central function for these proteins. Some move nucleosomes about on the genome in an ATP-dependent manner, some covalently modify histones such as methylating lysine 4 of histone H3, and some directly interact with the transcription machinery or are a part of that machinery. It is interesting to consider why these specific members of large families of functionally related proteins have strong developmental phenotypes.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures




Similar articles
-
The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila.Development. 2013 Oct;140(20):4182-92. doi: 10.1242/dev.095786. Epub 2013 Sep 4. Development. 2013. PMID: 24004944 Free PMC article.
-
Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27.Mol Cell Biol. 2012 Jun;32(12):2323-34. doi: 10.1128/MCB.06392-11. Epub 2012 Apr 9. Mol Cell Biol. 2012. PMID: 22493065 Free PMC article.
-
The Trithorax-mimic allele of Enhancer of zeste renders active domains of target genes accessible to polycomb-group-dependent silencing in Drosophila melanogaster.Genetics. 2001 Nov;159(3):1135-50. doi: 10.1093/genetics/159.3.1135. Genetics. 2001. PMID: 11729158 Free PMC article.
-
Genome regulation by polycomb and trithorax proteins.Cell. 2007 Feb 23;128(4):735-45. doi: 10.1016/j.cell.2007.02.009. Cell. 2007. PMID: 17320510 Review.
-
Noncoding RNAs in Polycomb and Trithorax Regulation: A Quantitative Perspective.Annu Rev Genet. 2017 Nov 27;51:385-411. doi: 10.1146/annurev-genet-120116-023402. Epub 2017 Sep 15. Annu Rev Genet. 2017. PMID: 28934594 Review.
Cited by
-
SMYD5 is a histone H3-specific methyltransferase mediating mono-methylation of histone H3 lysine 36 and 37.Biochem Biophys Res Commun. 2022 Apr 9;599:142-147. doi: 10.1016/j.bbrc.2022.02.043. Epub 2022 Feb 12. Biochem Biophys Res Commun. 2022. PMID: 35182940 Free PMC article.
-
Polycomb Assemblies Multitask to Regulate Transcription.Epigenomes. 2019 Jun 20;3(2):12. doi: 10.3390/epigenomes3020012. Epigenomes. 2019. PMID: 34968234 Free PMC article. Review.
-
Polycomb and Trithorax Group Proteins: The Long Road from Mutations in Drosophila to Use in Medicine.Acta Naturae. 2020 Oct-Dec;12(4):66-85. doi: 10.32607/actanaturae.11090. Acta Naturae. 2020. PMID: 33456979 Free PMC article.
-
A Genetic Mosaic Screen Reveals Ecdysone-Responsive Genes Regulating Drosophila Oogenesis.G3 (Bethesda). 2016 Aug 9;6(8):2629-42. doi: 10.1534/g3.116.028951. G3 (Bethesda). 2016. PMID: 27226164 Free PMC article.
-
The trxG protein ULT1 regulates Arabidopsis organ size by interacting with TCP14/15 to antagonize the LIM peptidase DA1 for H3K4me3 on target genes.Plant Commun. 2024 Apr 8;5(4):100819. doi: 10.1016/j.xplc.2024.100819. Epub 2024 Jan 12. Plant Commun. 2024. PMID: 38217289 Free PMC article.
References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103: 667–678 - PubMed
-
- Allis CD, Jenuwein T, Reinberg D 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
