The genetics of human skin disease
- PMID: 25274756
- PMCID: PMC4200211
- DOI: 10.1101/cshperspect.a015172
The genetics of human skin disease
Abstract
The skin is composed of a variety of cell types expressing specific molecules and possessing different properties that facilitate the complex interactions and intercellular communication essential for maintaining the structural integrity of the skin. Importantly, a single mutation in one of these molecules can disrupt the entire organization and function of these essential networks, leading to cell separation, blistering, and other striking phenotypes observed in inherited skin diseases. Over the past several decades, the genetic basis of many monogenic skin diseases has been elucidated using classical genetic techniques. Importantly, the findings from these studies has shed light onto the many classes of molecules and essential genetic as well as molecular interactions that lend the skin its rigid, yet flexible properties. With the advent of the human genome project, next-generation sequencing techniques, as well as several other recently developed methods, tremendous progress has been made in dissecting the genetic architecture of complex, non-Mendelian skin diseases.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
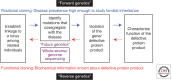
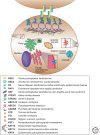

References
-
- Aberdam D, Galliano MF, Vailly J, Pulkkinen L, Bonifas J, Christiano AM, Tryggvason K, Uitto J, Epstein EH Jr, Ortonne JP, et al. 1994. Herlitz’s junctional epidermolysis bullosa is linked to mutations in the gene (LAMC2) for the γ2 subunit of nicein/kalinin (LAMININ-5). Nat Genet 6: 299–304 - PubMed
-
- Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA. 2003. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res 16: 208–214 - PubMed
-
- Almaani N, Nagy N, Liu L, Dopping-Hepenstal PJ, Lai-Cheong JE, Clements SE, Techanukul T, Tanaka A, Mellerio JE, McGrath JA. 2010. Revertant mosaicism in recessive dystrophic epidermolysis bullosa. J Invest Dermatol 130: 1937–1940 - PubMed
-
- Anton-Lamprecht I. 1983. Genetically induced abnormalities of epidermal differentiation and ultrastructure in ichthyoses and epidermolyses: Pathogenesis, heterogeneity, fetal manifestation, and prenatal diagnosis. J Invest Dermatol 81: 149s–156s - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
