Genetic labeling reveals novel cellular targets of schizophrenia susceptibility gene: distribution of GABA and non-GABA ErbB4-positive cells in adult mouse brain
- PMID: 25274830
- PMCID: PMC4180480
- DOI: 10.1523/JNEUROSCI.2021-14.2014
Genetic labeling reveals novel cellular targets of schizophrenia susceptibility gene: distribution of GABA and non-GABA ErbB4-positive cells in adult mouse brain
Abstract
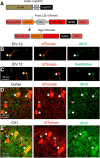
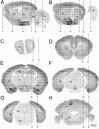
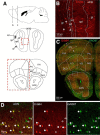
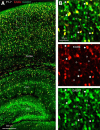
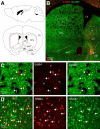
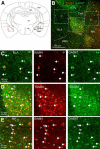
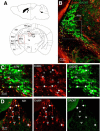
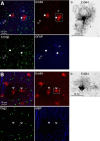
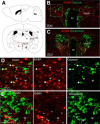
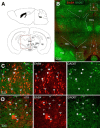
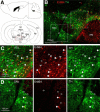
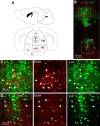
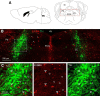
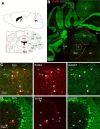
Neuregulin 1 (NRG1) and its receptor ErbB4 are schizophrenia risk genes. NRG1-ErbB4 signaling plays a critical role in neural development and regulates neurotransmission and synaptic plasticity. Nevertheless, its cellular targets remain controversial. ErbB4 was thought to express in excitatory neurons, although recent studies disputed this view. Using mice that express a fluorescent protein under the promoter of the ErbB4 gene, we determined in what cells ErbB4 is expressed and their identity. ErbB4 was widely expressed in the mouse brain, being highest in amygdala and cortex. Almost all ErbB4-positive cells were GABAergic in cortex, hippocampus, basal ganglia, and most of amygdala in neonatal and adult mice, suggesting GABAergic transmission as a major target of NRG1-ErbB4 signaling in these regions. Non-GABAergic, ErbB4-positive cells were present in thalamus, hypothalamus, midbrain, and hindbrain. In particular, ErbB4 is expressed in serotoninergic neurons of raphe nuclei but not in norepinephrinergic neurons of the locus ceruleus. In hypothalamus, ErbB4 is present in neurons that express oxytocin. Finally, ErbB4 is expressed in a group of cells in the subcortical areas that are positive for S100 calcium binding protein β. These results identify novel cellular targets of NRG1-ErbB4 signaling.
Keywords: ErbB4; NRG1; Neuregulin; S100β; oxytocin; serotonin.
Copyright © 2014 the authors 0270-6474/14/3413549-18$15.00/0.
Figures




References
-
- Barros CS, Calabrese B, Chamero P, Roberts AJ, Korzus E, Lloyd K, Stowers L, Mayford M, Halpain S, Müller U. Impaired maturation of dendritic spines without disorganization of cortical cell layers in mice lacking NRG1/ErbB signaling in the central nervous system. Proc Natl Acad Sci U S A. 2009;106:4507–4512. doi: 10.1073/pnas.0900355106. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
