Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage
- PMID: 25275497
- PMCID: PMC4183563
- DOI: 10.1371/journal.pone.0109213
Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage
Abstract
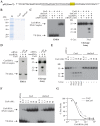
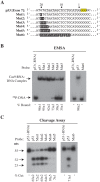
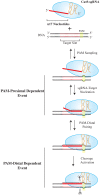
The clustered regularly interspaced short palindromic repeat (CRISPR)-associated enzyme Cas9 is an RNA-guided nuclease that has been widely adapted for genome editing in eukaryotic cells. However, the in vivo target specificity of Cas9 is poorly understood and most studies rely on in silico predictions to define the potential off-target editing spectrum. Using chromatin immunoprecipitation followed by sequencing (ChIP-seq), we delineate the genome-wide binding panorama of catalytically inactive Cas9 directed by two different single guide (sg) RNAs targeting the Trp53 locus. Cas9:sgRNA complexes are able to load onto multiple sites with short seed regions adjacent to (5')NGG(3') protospacer adjacent motifs (PAM). Yet among 43 ChIP-seq sites harboring seed regions analyzed for mutational status, we find editing only at the intended on-target locus and one off-target site. In vitro analysis of target site recognition revealed that interactions between the 5' end of the guide and PAM-distal target sequences are necessary to efficiently engage Cas9 nucleolytic activity, providing an explanation for why off-target editing is significantly lower than expected from ChIP-seq data.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

