SNP array profiling of mouse cell lines identifies their strains of origin and reveals cross-contamination and widespread aneuploidy
- PMID: 25277546
- PMCID: PMC4198738
- DOI: 10.1186/1471-2164-15-847
SNP array profiling of mouse cell lines identifies their strains of origin and reveals cross-contamination and widespread aneuploidy
Abstract
Background: The crisis of Misidentified and contaminated cell lines have plagued the biological research community for decades. Some repositories and journals have heeded calls for mandatory authentication of human cell lines, yet misidentification of mouse cell lines has received little publicity despite their importance in sponsored research. Short tandem repeat (STR) profiling is the standard authentication method, but it may fail to distinguish cell lines derived from the same inbred strain of mice. Additionally, STR profiling does not reveal karyotypic changes that occur in some high-passage lines and may have functional consequences. Single nucleotide polymorphism (SNP) profiling has been suggested as a more accurate and versatile alternative to STR profiling; however, a high-throughput method for SNP-based authentication of mouse cell lines has not been described.
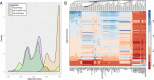
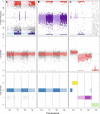
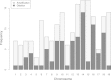
Results: We have developed computational methods (Cell Line Authentication by SNP Profiling, CLASP) for cell line authentication and copy number analysis based on a cost-efficient SNP array, and we provide a reference database of commonly used mouse strains and cell lines. We show that CLASP readily discriminates among cell lines of diverse taxonomic origins, including multiple cell lines derived from a single inbred strain, intercross or wild caught mouse. CLASP is also capable of detecting contaminants present at concentrations as low as 5%. Of the 99 cell lines we tested, 15 exhibited substantial divergence from the reported genetic background. In all cases, we were able to distinguish whether the authentication failure was due to misidentification (one cell line, Ba/F3), the presence of multiple strain backgrounds (five cell lines), contamination by other cells and/or the presence of aneuploid chromosomes (nine cell lines).
Conclusions: Misidentification and contamination of mouse cell lines is potentially as widespread as it is in human cell culture. This may have substantial implications for studies that are dependent on the expected background of their cell cultures. Laboratories can mitigate these risks by regular authentication of their cell cultures. Our results demonstrate that SNP array profiling is an effective method to combat cell line misidentification.
Figures

References
-
- Nature Identity crisis. Nature. 2009;457:935–936. - PubMed
-
- Podolak E: Ending Cell Line Contamination by Cutting off Researchers, Biotechniques Online News (2010).
-
- Yoshino K, Saijo K, Noro C, Nakamura Y. Development of a simple method to determine the mouse strain from which cultured cell lines originated. Interdisciplinary Bio Central. 2010;2:1–9. doi: 10.4051/ibc.2010.2.4.0014. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

