Structure of the large ribosomal subunit from human mitochondria
- PMID: 25278503
- PMCID: PMC4246062
- DOI: 10.1126/science.1258026
Structure of the large ribosomal subunit from human mitochondria
Abstract
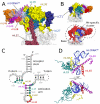
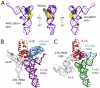
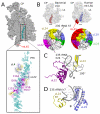
Human mitochondrial ribosomes are highly divergent from all other known ribosomes and are specialized to exclusively translate membrane proteins. They are linked with hereditary mitochondrial diseases and are often the unintended targets of various clinically useful antibiotics. Using single-particle cryogenic electron microscopy, we have determined the structure of its large subunit to 3.4 angstrom resolution, revealing 48 proteins, 21 of which are specific to mitochondria. The structure unveils an adaptation of the exit tunnel for hydrophobic nascent peptides, extensive remodeling of the central protuberance, including recruitment of mitochondrial valine transfer RNA (tRNA(Val)) to play an integral structural role, and changes in the tRNA binding sites related to the unusual characteristics of mitochondrial tRNAs.
Copyright © 2014, American Association for the Advancement of Science.
Figures



References
-
- O’Brien T. Properties of Human Mitochondrial Ribosomes. IUBMB Life. 2003;55:505–513. - PubMed
-
- Liu M, Spremulli L. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J Bio Chem. 2000;275:29400–29406. - PubMed
-
- Skladal D. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain. 2003;126:1905–1912. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

