Clinical microbiology informatics
- PMID: 25278581
- PMCID: PMC4187636
- DOI: 10.1128/CMR.00049-14
Clinical microbiology informatics
Abstract
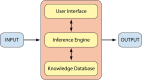
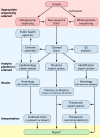
The clinical microbiology laboratory has responsibilities ranging from characterizing the causative agent in a patient's infection to helping detect global disease outbreaks. All of these processes are increasingly becoming partnered more intimately with informatics. Effective application of informatics tools can increase the accuracy, timeliness, and completeness of microbiology testing while decreasing the laboratory workload, which can lead to optimized laboratory workflow and decreased costs. Informatics is poised to be increasingly relevant in clinical microbiology, with the advent of total laboratory automation, complex instrument interfaces, electronic health records, clinical decision support tools, and the clinical implementation of microbial genome sequencing. This review discusses the diverse informatics aspects that are relevant to the clinical microbiology laboratory, including the following: the microbiology laboratory information system, decision support tools, expert systems, instrument interfaces, total laboratory automation, telemicrobiology, automated image analysis, nucleic acid sequence databases, electronic reporting of infectious agents to public health agencies, and disease outbreak surveillance. The breadth and utility of informatics tools used in clinical microbiology have made them indispensable to contemporary clinical and laboratory practice. Continued advances in technology and development of these informatics tools will further improve patient and public health care in the future.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures







References
-
- Campos JM. 2003. Clinical microbiology informatics—its time has come. Rev. Med. Microbiol. 14:105–108. 10.1097/00013542-200310000-00001 - DOI
-
- Yu VL, Fagan LM, Wraith SM, Clancey WJ, Scott AC, Hannigan J, Blum RL, Buchanan BG, Cohen SN. 1979. Antimicrobial selection by a computer. A blinded evaluation by infectious diseases experts. JAMA 242:1279–1282 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

