Selective microbial genomic DNA isolation using restriction endonucleases
- PMID: 25279840
- PMCID: PMC4184833
- DOI: 10.1371/journal.pone.0109061
Selective microbial genomic DNA isolation using restriction endonucleases
Abstract
To improve the metagenomic analysis of complex microbiomes, we have repurposed restriction endonucleases as methyl specific DNA binding proteins. As an example, we use DpnI immobilized on magnetic beads. The ten minute extraction technique allows specific binding of genomes containing the DpnI Gm6ATC motif common in the genomic DNA of many bacteria including γ-proteobacteria. Using synthetic genome mixtures, we demonstrate 80% recovery of Escherichia coli genomic DNA even when only femtogram quantities are spiked into 10 µg of human DNA background. Binding is very specific with less than 0.5% of human DNA bound. Next Generation Sequencing of input and enriched synthetic mixtures results in over 100-fold enrichment of target genomes relative to human and plant DNA. We also show comparable enrichment when sequencing complex microbiomes such as those from creek water and human saliva. The technique can be broadened to other restriction enzymes allowing for the selective enrichment of trace and unculturable organisms from complex microbiomes and the stratification of organisms according to restriction enzyme enrichment.
Conflict of interest statement
Figures
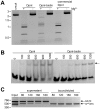
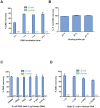

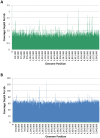
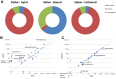

References
-
- Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11: 789–799. - PubMed
-
- Engel P, Moran NA (2013) The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev 37: 699–735. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

