Evolutionary strategies of viruses, bacteria and archaea in hydrothermal vent ecosystems revealed through metagenomics
- PMID: 25279954
- PMCID: PMC4184897
- DOI: 10.1371/journal.pone.0109696
Evolutionary strategies of viruses, bacteria and archaea in hydrothermal vent ecosystems revealed through metagenomics
Abstract
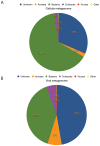
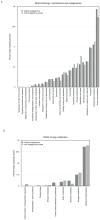
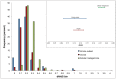
The deep-sea hydrothermal vent habitat hosts a diverse community of archaea and bacteria that withstand extreme fluctuations in environmental conditions. Abundant viruses in these systems, a high proportion of which are lysogenic, must also withstand these environmental extremes. Here, we explore the evolutionary strategies of both microorganisms and viruses in hydrothermal systems through comparative analysis of a cellular and viral metagenome, collected by size fractionation of high temperature fluids from a diffuse flow hydrothermal vent. We detected a high enrichment of mobile elements and proviruses in the cellular fraction relative to microorganisms in other environments. We observed a relatively high abundance of genes related to energy metabolism as well as cofactors and vitamins in the viral fraction compared to the cellular fraction, which suggest encoding of auxiliary metabolic genes on viral genomes. Moreover, the observation of stronger purifying selection in the viral versus cellular gene pool suggests viral strategies that promote prolonged host integration. Our results demonstrate that there is great potential for hydrothermal vent viruses to integrate into hosts, facilitate horizontal gene transfer, and express or transfer genes that manipulate the hosts' functional capabilities.
Conflict of interest statement
Figures


References
-
- Anderson RE, Brazelton WJ, Baross JA (2013) The deep viriosphere: Assessing the viral impact on microbial community dynamics in the deep subsurface. Rev Mineral Geochemistry 75: 649–675.
-
- Anderson RE, Brazelton WJ, Baross JA (2011) Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol Ecol 77: 120–133. - PubMed
-
- Baross JA, Hoffman SE (1985) Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Orig Life Evol Biosph 15: 327–345.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

