Effective genetic-risk prediction using mixed models
- PMID: 25279982
- PMCID: PMC4185122
- DOI: 10.1016/j.ajhg.2014.09.007
Effective genetic-risk prediction using mixed models
Abstract
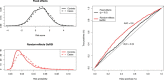
For predicting genetic risk, we propose a statistical approach that is specifically adapted to dealing with the challenges imposed by disease phenotypes and case-control sampling. Our approach (termed Genetic Risk Scores Inference [GeRSI]), combines the power of fixed-effects models (which estimate and aggregate the effects of single SNPs) and random-effects models (which rely primarily on whole-genome similarities between individuals) within the framework of the widely used liability-threshold model. We demonstrate in extensive simulation that GeRSI produces predictions that are consistently superior to current state-of-the-art approaches. When applying GeRSI to seven phenotypes from the Wellcome Trust Case Control Consortium (WTCCC) study, we confirm that the use of random effects is most beneficial for diseases that are known to be highly polygenic: hypertension (HT) and bipolar disorder (BD). For HT, there are no significant associations in the WTCCC data. The fixed-effects model yields an area under the ROC curve (AUC) of 54%, whereas GeRSI improves it to 59%. For BD, using GeRSI improves the AUC from 55% to 62%. For individuals ranked at the top 10% of BD risk predictions, using GeRSI substantially increases the BD relative risk from 1.4 to 2.5.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Goldstein D.B. Common genetic variation and human traits. N. Engl. J. Med. 2009;360:1696–1698. - PubMed
-
- Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O’Donovan M.C., Sullivan P.F., Sklar P., Ruderfer D.M., McQuillin A., Morris D.W., International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

