Characteristics of neutral and deleterious protein-coding variation among individuals and populations
- PMID: 25279984
- PMCID: PMC4185119
- DOI: 10.1016/j.ajhg.2014.09.006
Characteristics of neutral and deleterious protein-coding variation among individuals and populations
Abstract
Whole-genome and exome data sets continue to be produced at a frenetic pace, resulting in massively large catalogs of human genomic variation. However, a clear picture of the characteristics and patterns of neutral and deleterious variation within and between populations has yet to emerge, given that recent large-scale sequencing studies have often emphasized different aspects of the data and sometimes appear to have conflicting conclusions. Here, we comprehensively studied characteristics of protein-coding variation in high-coverage exome sequence data from 6,515 European American (EA) and African American (AA) individuals. We developed an unbiased approach to identify putatively deleterious variants and investigated patterns of neutral and deleterious single-nucleotide variants and alleles between individuals and populations. We show that there are substantial differences in the composition of genotypes between EA and AA populations and that small but statistically significant differences exist in the average number of deleterious alleles carried by EA and AA individuals. Furthermore, we performed extensive simulations to delineate the temporal dynamics of deleterious alleles for a broad range of demographic models and use these data to inform the interpretation of empirical patterns of deleterious variation. Finally, we illustrate that the effects of demographic perturbations, such as bottlenecks and expansions, often manifest in opposing patterns of neutral and deleterious variation depending on whether the focus is on populations or individuals. Our results clarify seemingly disparate empirical characteristics of protein-coding variation and provide substantial insights into how natural selection and demographic history have patterned neutral and deleterious variation within and between populations.
Copyright © 2014 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
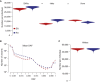

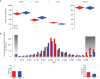

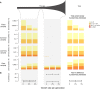
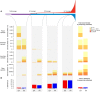
Comment in
-
Human genetics: Dissecting deleterious mutation complexity.Nat Rev Genet. 2014 Dec;15(12):777. doi: 10.1038/nrg3852. Epub 2014 Oct 29. Nat Rev Genet. 2014. PMID: 25352302 No abstract available.
References
-
- Haldane J.B.S. The effect of variation on fitness. Am. Nat. 1937;71:337–349.
-
- Crow J.F. Genetic Loads and the Cost of Natural Selection. In: Kojima K.-i., editor. Mathematical Topics in Population Genetics. Springer-Verlag; New York: 1970. pp. 128–177.
-
- Crow J.F. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 2000;1:40–47. - PubMed
-
- Gavrilov L.A., Gavrilova N.S., Kroutko V.N., Evdokushkina G.N., Semyonova V.G., Gavrilova A.L., Lapshin E.V., Evdokushkina N.N., Kushnareva Y.E. Mutation load and human longevity. Mutat. Res. 1997;377:61–62. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

