Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes
- PMID: 25280220
- PMCID: PMC4201500
- DOI: 10.1016/j.stem.2014.09.002
Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes
Abstract
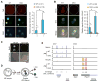
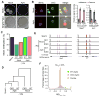
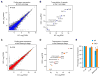
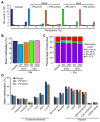
With the exception of imprinted genes and certain repeats, DNA methylation is globally erased during preimplantation development. Recent studies have suggested that Tet3-mediated oxidation of 5-methylcytosine (5mC) and DNA replication-dependent dilution both contribute to global paternal DNA demethylation, but demethylation of the maternal genome occurs via replication. Here we present genome-scale DNA methylation maps for both the paternal and maternal genomes of Tet3-depleted and/or DNA replication-inhibited zygotes. In both genomes, we found that inhibition of DNA replication blocks DNA demethylation independently from Tet3 function and that Tet3 facilitates DNA demethylation largely by coupling with DNA replication. For both genomes, our data indicate that replication-dependent dilution is the major contributor to demethylation, but Tet3 plays an important role, particularly at certain loci. Our study thus defines the respective functions of Tet3 and DNA replication in paternal DNA demethylation and reveals an unexpected contribution of Tet3 to demethylation of the maternal genome.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures


References
-
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. - PubMed
-
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. - PubMed
-
- Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, Yan J, Ren X, Lin S, Li J, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

