Meta-analysis of diabetic nephropathy associated genetic variants in inflammation and angiogenesis involved in different biochemical pathways
- PMID: 25280384
- PMCID: PMC4411872
- DOI: 10.1186/s12881-014-0103-8
Meta-analysis of diabetic nephropathy associated genetic variants in inflammation and angiogenesis involved in different biochemical pathways
Abstract
Background: Diabetes mellitus is the most common chronic endocrine disorder, affecting an estimated population of 382 million people worldwide. It is associated with microvascular and macrovascular complications, including diabetic nephropathy (DN); primary cause of end-stage renal disease. Different inflammatory and angiogenic molecules in various pathways are important modulators in the pathogenesis and progression of diabetic nephropathy. Differential disease risk in DN may be partly attributable to genetic susceptibility. In this meta-analysis, we aimed to determine which of the previously investigated genetic variants in these pathways are significantly associated with the development of DN and to examine the functional role of these genes.
Methods: A systematic search was conducted to collect and analyze all studies published till June 2013; that investigated the association between genetic variants involved in inflammatory cytokines and angiogenesis and diabetic nephropathy. Genetic variants associated with DN were selected and analyzed by using Comprehensive Meta Analysis software. Pathway analysis of the genes with variants showing significant positive association with DN was performed using Genomatix Genome Analyzer (Genomatix, Munich, Germany).
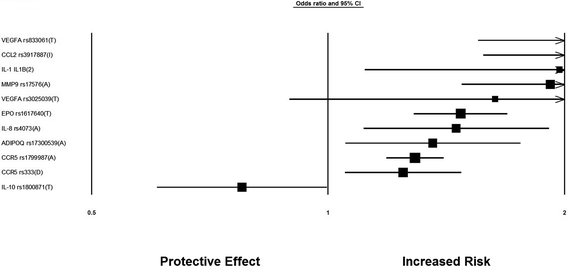
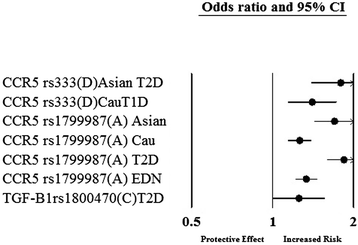
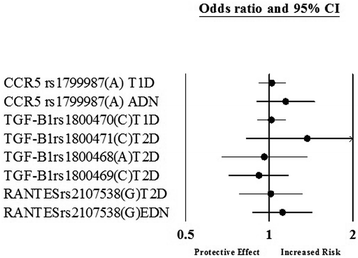
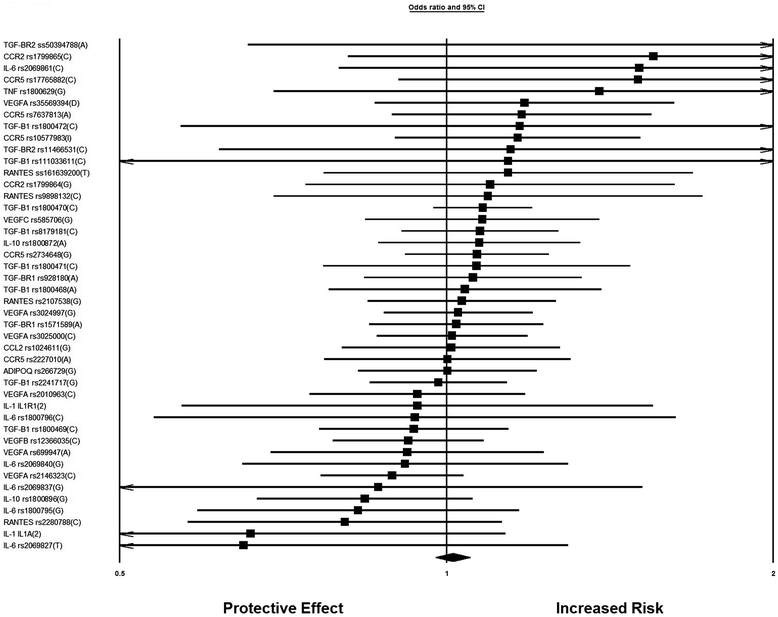
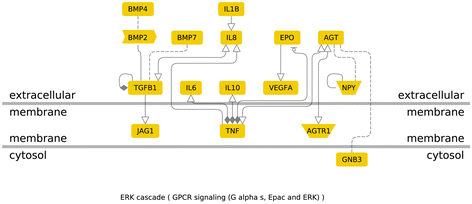
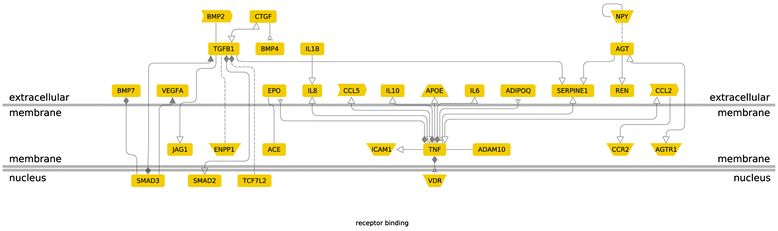
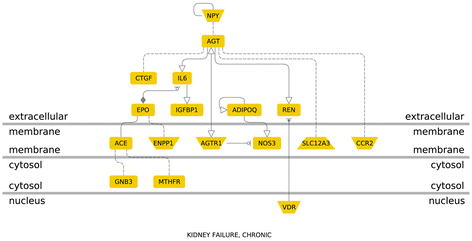
Results: After the inclusion and exclusion criteria for this analysis, 34 studies were included in this meta-analysis. 11 genetic variants showed significant positive association with DN in a random-effects meta-analysis. These included genetic variants within or near VEGFA, CCR5, CCL2, IL-1, MMP9, EPO, IL-8, ADIPOQ and IL-10. rs1800871 (T) genetic variant in IL-10 showed protective effect for DN. Most of these eleven genetic variants were involved in GPCR signaling and receptor binding pathways whereas four were involved in chronic kidney failure. rs833061 [OR 2.08 (95% CI 1.63-2.66)] in the VEGFA gene and rs3917887 [OR 2.04 (95% CI 1.64-2.54)] in the CCL2 gene showed the most significant association with the risk of diabetic nephropathy.
Conclusions: Our results indicate that 11 genetic variants within or near VEGFA, CCR5, CCL2, IL-1, MMP9, EPO, IL-8, ADIPOQ and IL-10 showed significant positive association with diabetic nephropathy. Gene Ontology or pathway analysis showed that these genes may contribute to the pathophysiology of DN. The functional relevance of the variants and their pathways can lead to increased biological insights and development of new therapeutic targets.
Figures
Similar articles
-
Association of eNOS and MCP-1 Genetic Variants with Type 2 Diabetes and Diabetic Nephropathy Susceptibility: A Case-Control and Meta-Analysis Study.Biochem Genet. 2021 Aug;59(4):966-996. doi: 10.1007/s10528-021-10041-2. Epub 2021 Feb 20. Biochem Genet. 2021. PMID: 33609191 Free PMC article.
-
Common variants of inflammatory cytokine genes are associated with risk of nephropathy in type 2 diabetes among Asian Indians.PLoS One. 2009;4(4):e5168. doi: 10.1371/journal.pone.0005168. Epub 2009 Apr 9. PLoS One. 2009. PMID: 19357773 Free PMC article.
-
Association of monocyte chemoattractant protein-1 2518G/A gene polymorphism with diabetic nephropathy risk.J Recept Signal Transduct Res. 2015 Feb;35(1):94-7. doi: 10.3109/10799893.2014.936458. Epub 2014 Jul 22. J Recept Signal Transduct Res. 2015. PMID: 25051493
-
Association between polymorphisms of cytokine genes and diabetic nephropathy: A comprehensive systematic review and meta-analysis.Int J Clin Pract. 2021 Nov;75(11):e14634. doi: 10.1111/ijcp.14634. Epub 2021 Aug 8. Int J Clin Pract. 2021. PMID: 34309136
-
The genetic map of diabetic nephropathy: evidence from a systematic review and meta-analysis of genetic association studies.Clin Kidney J. 2020 Jul 12;13(5):768-781. doi: 10.1093/ckj/sfaa077. eCollection 2020 Oct. Clin Kidney J. 2020. PMID: 33123356 Free PMC article. Review.
Cited by
-
Diabetic kidney disease in type 2 diabetes: a review of pathogenic mechanisms, patient-related factors and therapeutic options.PeerJ. 2021 Apr 19;9:e11070. doi: 10.7717/peerj.11070. eCollection 2021. PeerJ. 2021. PMID: 33976959 Free PMC article.
-
Common Inflammation-Related Candidate Gene Variants and Acute Kidney Injury in 2647 Critically Ill Finnish Patients.J Clin Med. 2019 Mar 11;8(3):342. doi: 10.3390/jcm8030342. J Clin Med. 2019. PMID: 30862128 Free PMC article.
-
Association of CCL2, CCR5, ELMO1, and IL8 Polymorphism with Diabetic Nephropathy in Malaysian Type 2 Diabetic Patients.Int J Chronic Dis. 2019 Jan 1;2019:2053015. doi: 10.1155/2019/2053015. eCollection 2019. Int J Chronic Dis. 2019. PMID: 30713847 Free PMC article.
-
Association of chemokine ligand 5/chemokine receptor 5 gene promoter polymorphisms with diabetic microvascular complications: A meta-analysis.J Diabetes Investig. 2016 Mar;7(2):212-8. doi: 10.1111/jdi.12397. Epub 2015 Aug 21. J Diabetes Investig. 2016. PMID: 27042273 Free PMC article.
-
Aortic Stiffness Index And Carotid Intima-Media Thickness Are Independently Associated With The Presence Of Microalbuminuria In Patients With Type 2 Diabetes Mellitus.Diabetes Metab Syndr Obes. 2019 Sep 19;12:1889-1896. doi: 10.2147/DMSO.S223880. eCollection 2019. Diabetes Metab Syndr Obes. 2019. PMID: 31571963 Free PMC article.
References
-
- International Diabetes Federation: IDF Diabetes Atlas. 6th edition. Brussels, Belgium: International Diabetes Federation; 2013. [http://www.idf.org/diabetesatlas]
-
- Ezzidi I, Mtiraoui N, Kacem M, Mallat SG, Mohamed MB, Chaieb M, Mahjoub T, Almawi WY. Interleukin-10–592C/A, −819C/T and –1082A/G promoter variants affect the susceptibility to nephropathy in Tunisian type 2 diabetes (T2DM) patients. Clin Endocrinol. 2009;70:401–407. doi: 10.1111/j.1365-2265.2008.03337.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

