Transcriptome analysis of Gossypium hirsutum flower buds infested by cotton boll weevil (Anthonomus grandis) larvae
- PMID: 25280771
- PMCID: PMC4234063
- DOI: 10.1186/1471-2164-15-854
Transcriptome analysis of Gossypium hirsutum flower buds infested by cotton boll weevil (Anthonomus grandis) larvae
Abstract
Background: Cotton is a major fibre crop grown worldwide that suffers extensive damage from chewing insects, including the cotton boll weevil larvae (Anthonomus grandis). Transcriptome analysis was performed to understand the molecular interactions between Gossypium hirsutum L. and cotton boll weevil larvae. The Illumina HiSeq 2000 platform was used to sequence the transcriptome of cotton flower buds infested with boll weevil larvae.
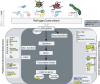
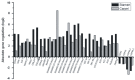
Results: The analysis generated a total of 327,489,418 sequence reads that were aligned to the G. hirsutum reference transcriptome. The total number of expressed genes was over 21,697 per sample with an average length of 1,063 bp. The DEGseq analysis identified 443 differentially expressed genes (DEG) in cotton flower buds infected with boll weevil larvae. Among them, 402 (90.7%) were up-regulated, 41 (9.3%) were down-regulated and 432 (97.5%) were identified as orthologues of A. thaliana genes using Blastx. Mapman analysis of DEG indicated that many genes were involved in the biotic stress response spanning a range of functions, from a gene encoding a receptor-like kinase to genes involved in triggering defensive responses such as MAPK, transcription factors (WRKY and ERF) and signalling by ethylene (ET) and jasmonic acid (JA) hormones. Furthermore, the spatial expression pattern of 32 of the genes responsive to boll weevil larvae feeding was determined by "in situ" qPCR analysis from RNA isolated from two flower structures, the stamen and the carpel, by laser microdissection (LMD).
Conclusion: A large number of cotton transcripts were significantly altered upon infestation by larvae. Among the changes in gene expression, we highlighted the transcription of receptors/sensors that recognise chitin or insect oral secretions; the altered regulation of transcripts encoding enzymes related to kinase cascades, transcription factors, Ca2+ influxes, and reactive oxygen species; and the modulation of transcripts encoding enzymes from phytohormone signalling pathways. These data will aid in the selection of target genes to genetically engineer cotton to control the cotton boll weevil.
Figures



References
-
- Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. doi: 10.1016/S0065-2113(02)78004-8. - DOI
-
- Razaq M, Aslam M, Shad SA, Naeem M. Evaluation of some new promising cotton strains against bollworm complex. Evaluation. 2004;15(3):313–318.
-
- Dubey NK, Goel R, Ranjan A, Idris A, Singh SK, Bag SK, Chandrashekar K, Pandey KD, Singh PK, Sawant SV. Comparative transcriptome analysis of Gossypium hirsutum L. in response to sap sucking insects: aphid and whitefly. BMC Genomics. 2013;14:241–261. doi: 10.1186/1471-2164-14-241. - DOI - PMC - PubMed
-
- Greenberg SM, Sappington TW, Setamou M, Coleman RJ. Influence of different cotton fruit sizes on boll weevil (Coleoptera: Curculionidae) oviposition and survival to adulthood. Environ Entomol. 2003;33:443–449. doi: 10.1603/0046-225X-33.2.443. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

